SidechainNet
SidechainNet is a protein structure prediction dataset that directly extends ProteinNet1 by Mohammed AlQuraishi.
Specifically, SidechainNet adds measurements for protein angles and coordinates that describe the complete, all-atom protein structure (backbone and sidechain, excluding hydrogens) instead of the protein backbone alone.
This repository provides the following:
- SidechainNet datasets stored as pickled Python dictionaries.
- Methods for loading and batching SidechainNet data efficiently in PyTorch.
- Methods for generating protein structure visualizations (
.pdb,3Dmol,.gltf) from model predictions.
Summary of SidechainNet data
| Entry | Dimensionality* | Label in SidechainNet data | ProteinNet | SidechainNet |
|---|---|---|---|---|
| Primary sequence | L x 1 | seq |
X | X |
| PSSM + Information content | L x 21 | evo |
X | X |
| Missing residue mask | L x 1 | msk |
X | X |
| Backbone coordinates | L x 4** x 3 | crd, subset [0:4] |
X | X |
| Backbone torsion angles | L x 3 | ang, subset [0:3] |
X | |
| Backbone bond angles | L x 3 | ang, subset [3:6] |
X | |
| Sidechain torsion angles | L x 6 | ang, subset [6:12] |
X | |
| Sidechain coordinates | L x 10 x 3 | crd, subset [4:14] |
X |
*L reperesents the length of any given protein in the dataset.
**SidechainNet explicitly includes oxygen atoms as part of the backbone coordinate data in contrast to ProteinNet, which only includes the primary N, C_alpha, C atoms.
Installation
To run this code, it's recommended to first clone the repo into an appropriate source directory with git clone <CLONE_URL>. Then, perform a developmental install of the package with pip in your current environment with pip install -e .. This will install the sidechainnet package in your environment.
Usage Examples
Loading SidechainNet as a Python dictionary
import sidechainnet as scn
data = scn.load(casp_version=12, thinning=30)In its most basic form, SidechainNet is stored as a Python dictionary organized by train, validation, and test splits. These splits are identical to those described in ProteinNet.
Within each train/validation/test split in SidechainNet is another dictionary mapping data entry types (seq, ang, etc.) to a list containing this data type for every protein. In the example below, seq{i}, ang{i}, ... all refer to the ith protein in the dataset.
data = {"train": {"seq": [seq1, seq2, ...], # Sequences
"ang": [ang1, ang2, ...], # Angles
"crd": [crd1, crd2, ...], # Coordinates
"evo": [evo1, evo2, ...], # PSSMs and Information Content
"ids": [id1, id2, ...], # Corresponding ProteinNet IDs
},
"valid-10": {...},
...
"valid-90": {...},
"test": {...},
"settings": {...},
"description" : "SidechainNet for CASP 12."
"date": "September 20, 2020"
}By default, the load function downloads the data from the web into the current directory and loads it as a Python dictionary. If the data already exists locally, it reads it from disk. Other than the requirement that the data must be loaded using Python, this method of data loading is agnostic to any downstream analyses
Loading SidechainNet with PyTorch DataLoaders
The load function can also be used to load SidechainNet data as a dictionary of torch.utils.data.DataLoader objects. PyTorch DataLoaders make it simple to iterate over dataset items for training machine learning models. This method is recommended for using SidechainNet data with PyTorch.
By default, the provided DataLoaders use a custom batching method that randomly generates batches of proteins of similar length. For faster training, it generates larger batches when the average length of proteins in the batch is small, and smaller batches when the proteins are large. The probability of selecting small-length batches is decreased so that each protein in SidechainNet is included in a batch with equal probability. See dynamic_batching and collate_fn arguments for more information on modifying this behavior. In the example below, model_input is a collated Tensor containing sequence and PSSM information.
>>> dataloaders = scn.load(casp_version=12, with_pytorch="dataloaders")
>>> dataloaders.keys()
['train', 'train_eval', 'valid-10', ..., 'valid-90', 'test']
>>> dataloaders['train'].dataset
ProteinDataset(casp_version=12, split='train', n_proteins=81454,
created='Sep 20, 2020')
>>> for protein_id, model_input, true_angles, true_coords in dataloaders['train']:
.... predicted_angles = model(model_input)
.... predicted_coords = angles_to_coordinates(predicted_angles)
.... loss = compute_loss(predicted_angles, predicted_coords,
true_angles, true_coords)
.... ...We have also made it possible to access the protein sequence, mask, and PSSM data directly when training by adding aggregate_model_input=False to scn.load.
>>> dataloaders = scn.load(casp_version=12, with_pytorch="dataloaders",
aggregate_model_input=False)
>>> for (protein_id, sequence, mask, pssm, true_angles,
true_coords) in dataloaders['train']:
.... prediction = model(sequence, pssm)
.... ...Converting Angle Representations into All-Atom Structures
An important component of this work is the inclusion of both angular and 3D coordinate representations of each protein. Researchers who develop methods that rely on angular representations may be interested in converting this information into 3D coordinates. For this reason, SidechainNet provides a method to convert the angles it provides into Cartesian coordinates.
In the below example, angles is a NumPy matrix or Torch Tensor following the same organization as the NumPy angle matrices provided in SidechainNet. sequence is a string representing the protein's amino acid sequence.
>>> (len(sequence), angles.shape) # 12 angles per residue
(128, (128, 12))
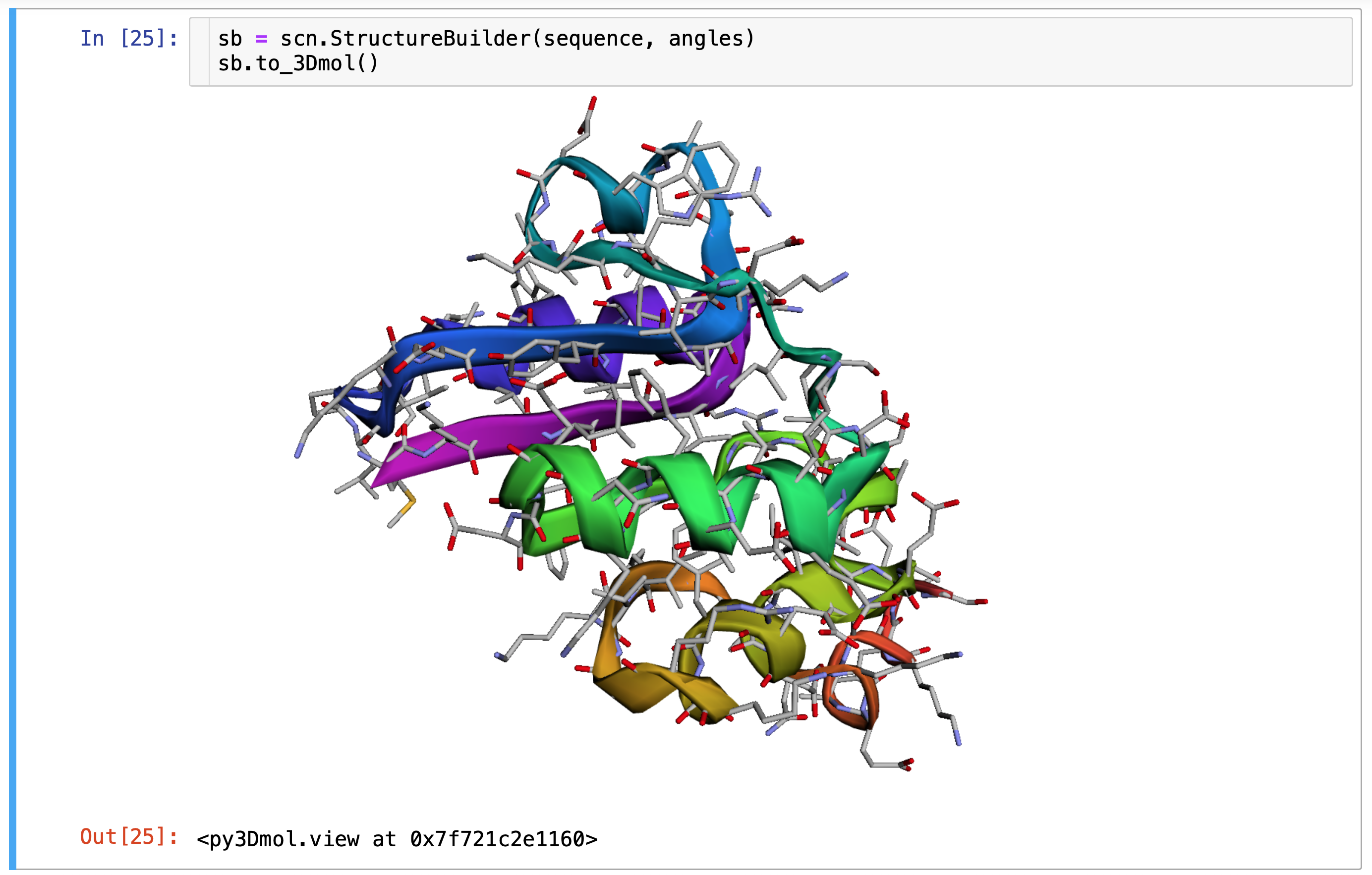
>>> sb = scn.StructureBuilder(sequence, angles)
>>> coords = sb.build()
>>> coords.shape # 14 atoms per residue (128*14 = 1792)
(1792, 3)Visualizing & Interacting with All-Atom Structures (PDB, py3Dmol, and gLTF)
SidechainNet also makes it easy to visualize both existing and predicted all-atom protein structures. These visualizations are available as PDB files, py3Dmol.view objects, and Graphics Library Transmission Format (gLTF) files. Examples of each are included below.
The PDB format is a typical format for representing protein structures and can be opened in software tools like PyMOL. py3Dmol (built on 3Dmol.js2) enables users to visualize and interact with protein structures on the web and in Jupyter Notebooks via an open-source, object-oriented, and hardware-accelerated Javascript library. Finally, gLTF files, despite their larger size, can be convenient for visualizing proteins on the web or in contexts where other protein visualization tools are not supported.
>>> sb.to_pdb("example.pdb")
>>> sb.to_gltf("example.gltf")Using SidechainNet to train an all-atom protein structure prediction model
Below is an outline of how to use this repository for machine learning model training. Assuming you have a predictive model variable model and a loss function loss_fn used to evaluate your model, you can load SidechainNet using our DataLoaders and begin training.
import sidechainnet as scn
data = scn.load(casp_version=12,thinning=30, with_pytorch="dataloaders")
for epoch in range(100):
# Training epoch
for protein_ids, model_input, tgt_angles, tgt_coords in data['train']:
predictions = model(model_input)
loss = loss_fn(predictions, tgt_angles, tgt_coords)
loss.backwards()
...
# Evaluate performance on down-sampled training set for efficiency
for protein_ids, model_input, tgt_angles, tgt_coords in data['train-eval']:
predictions = model(model_input)
loss = loss_fn(predictions, tgt_angles, tgt_coords)
loss.backwards()
...
# Evaluate performance on each of the 7 validation sets
for valid_set in [data[f'valid-{split}'] for split in scn.utils.download.VALID_SPLITS]:
for protein_ids, model_input, tgt_angles, tgt_coords in valid_set:
predictions = model(model_input)
loss = loss_fn(predictions, tgt_angles, tgt_coords)
loss.backwards()
...
# Evaluate performance on test set
for protein_ids, model_input, tgt_angles, tgt_coords in data['test']:
predictions = model(model_input)
loss = loss_fn(predictions, tgt_angles, tgt_coords)
...Reproducing SidechainNet
If you would like to reproduce our work or make modifications to the dataset, you may follow these directions below to generate SidechainNet from scratch.
Package Requirements
- Python 3
- ProDy (
pip install ProDy)- Biopython
- numpy
- scipy
- PyTorch
- tqdm
- py3Dmol (
pip install py3Dmol) - pymol (optional, for
gltfsupport, repo, linux install requireslibxml2)
Acknowledgements
Thanks to Mohammed AlQuraishi for his inspiring work on protein structure prediction. Thanks, also, to Jeppe Hallgren for his development of a ProteinNet text record parser, which I have used in part here.
This work is supported by R01GM108340 from the National Institute of General Medical Sciences, is supported in part by the University of Pittsburgh Center for Research Computing through the resources provided, and by NIH T32 training grant T32 EB009403 as part of the HHMI-NIBIB Interfaces Initiative.
Project structure (continuous integration, docs, testing) based on the Computational Molecular Science Python Cookiecutter version 1.1.
References
- ProteinNet: a standardized data set for machine learning of protein structure.. M. AlQuraishi. BMC Bioinformatics 20, 311 (2019).
- 3dmol.js: molecular visualization with WebGL. N. Rego and D. Koes. Bioinformatics, 31(8):1322–1324, (2014).
Copyright
Copyright (c) 2020, Jonathan King