Efficient implementation of an cell type annotation method
- ⚡ support multi-core processing
- ⚡ support spare matrix to save memory
- ⚡ pure python
Collection of cell type specific open chromatin regions to use
Tutorials
Command line usage
-
usage:
cli_run.py [-h] [-o OUTPUT] [-t THRESHOLD] [--cpu CPU] [--verbose] [--umap_fig] [-u UMAP_INPUT] [-f OUTPUT_FIG] count_input ref_folder
positional arguments:
- count_input path to 10x input folder
- ref_folder path to a folder of cell type specific peaks
optional arguments:
- -h, --help show this help message and exit
- -o OUTPUT, --output OUTPUT name of the output file
- -t THRESHOLD, --threshold THRESHOLD threshold to compute bed overlap. a float number ranges from 0 - 1.
- --cpu CPU number of cpus
- --verbose flag to print messages
- --umap_fig flag to skip ploting umap
- -u UMAP_INPUT, --umap_input UMAP_INPUT path to a csv file of umap coordinates (cell x 2). The file should have an index column of cell barcodes
- -f OUTPUT_FIG, --output_fig OUTPUT_FIG name of the output png figure
-
Example:
-
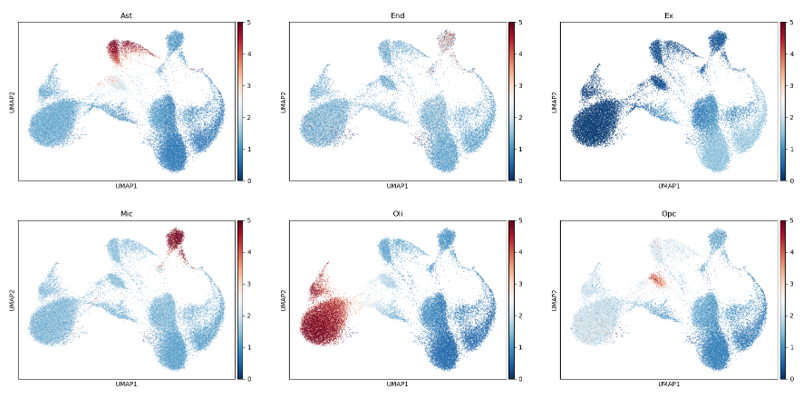
To generate the cover figure, run
python src/cli_run.py case_study/brain_atac/filtered_peak_bc_matrix/ OCR_DB/GSE97887/ --cpu 10
A file (score.csv) and png figure (score.png) will be outputted.
-