LucaOne: Generalized Biological Foundation Model with Unified Nucleic Acid and Protein Language.
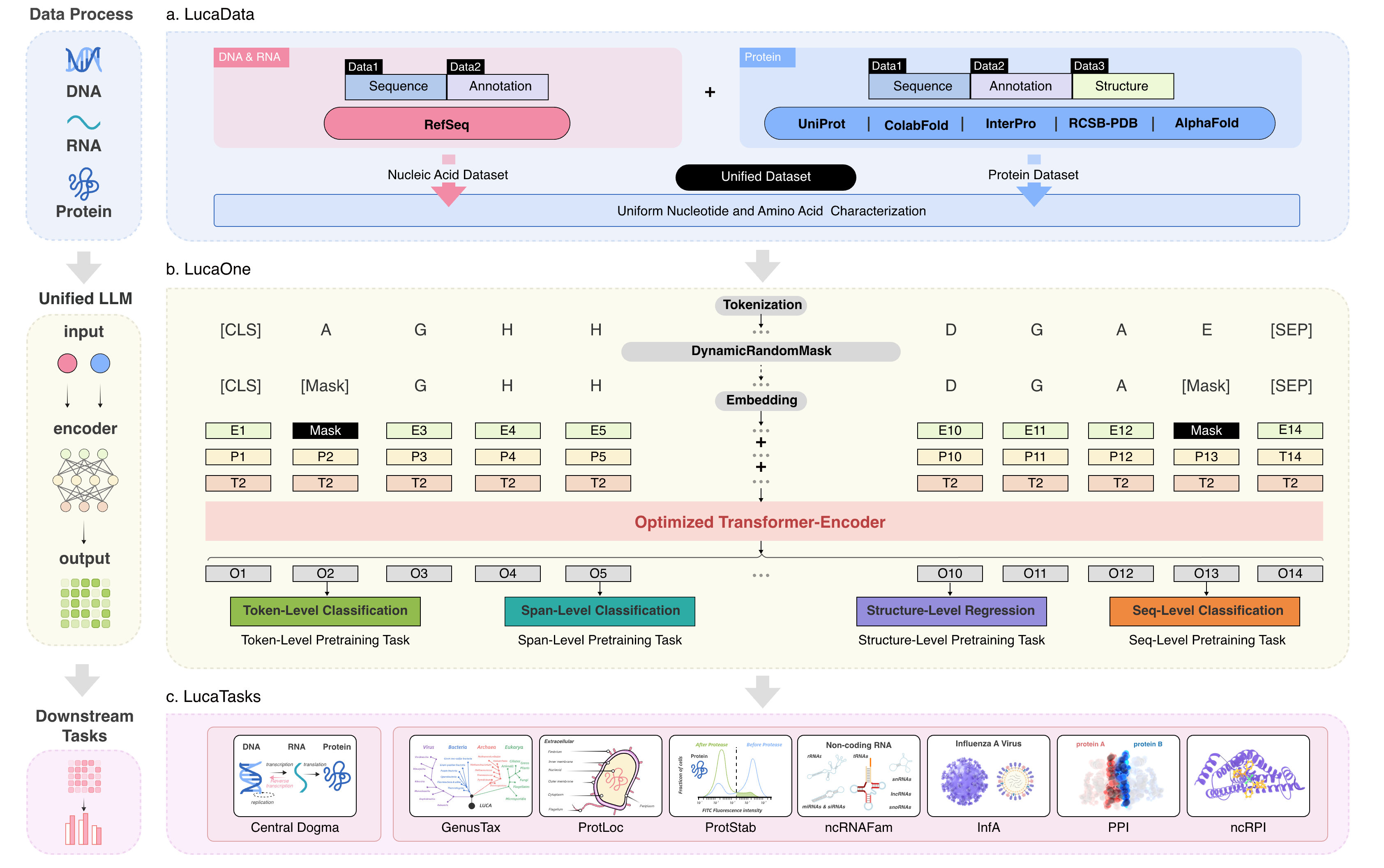
Fig. 1 The workflow of LucaOne.
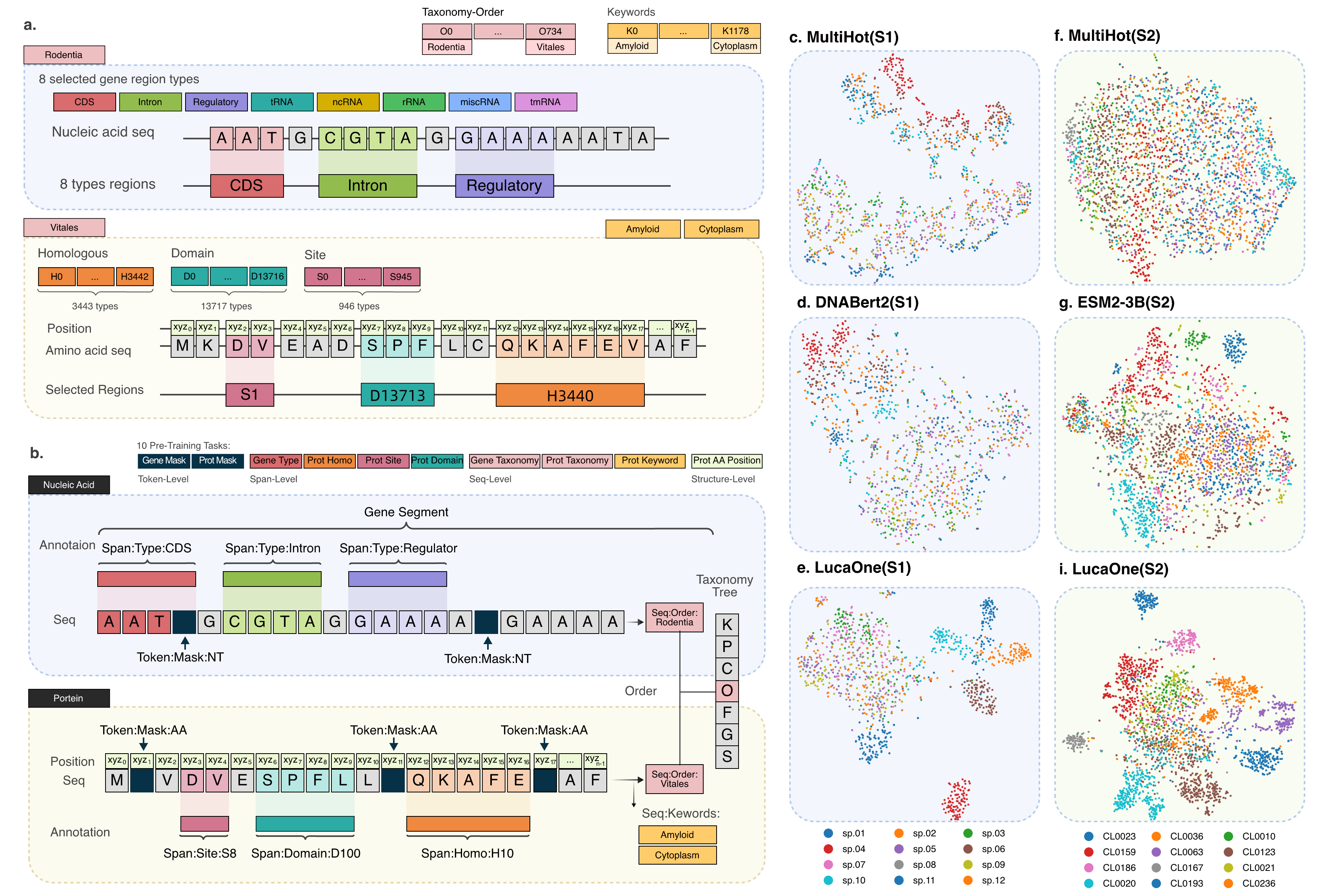
Fig. 2 The data and tasks for pre-training LucaOne, and T-SNE on four embedding models.
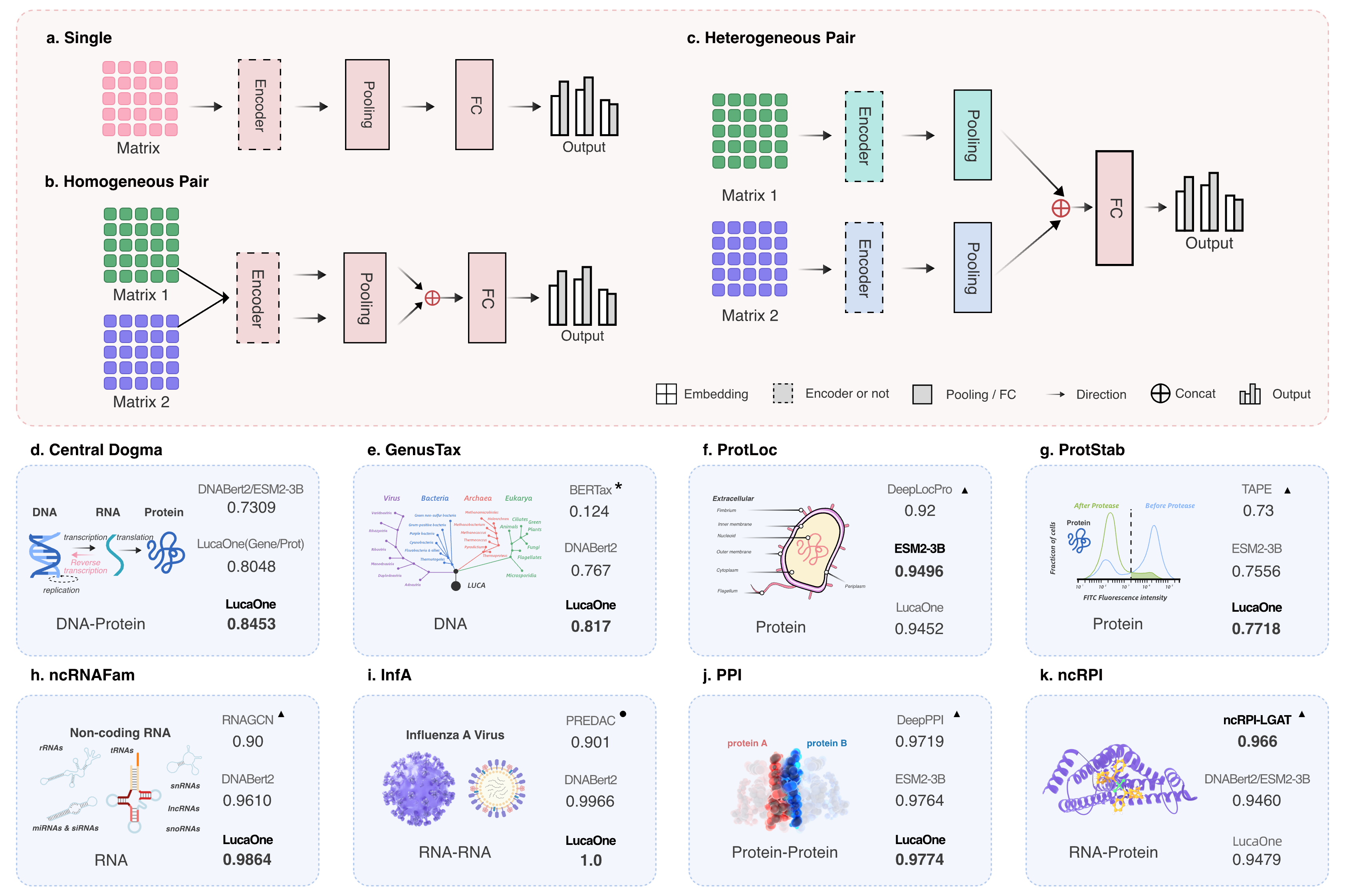
Fig. 3 Downstream task network with three input types and results comparison of 8 verification tasks.
sudo yum update
sudo yum install git-all
sudo apt-get update
sudo apt install git-all
wget https://repo.anaconda.com/archive/Anaconda3-2022.05-Linux-x86_64.sh
sh Anaconda3-2022.05-Linux-x86_64.sh
source ~/.bashrc
conda create -n lucaone python=3.9.13
conda activate lucaone
pip install -r requirements.txt -i https://pypi.tuna.tsinghua.edu.cn/simple
Pretraining Dataset FTP: Dataset for LucaOne
Copy the dataset from http://47.93.21.181/lucaone/PreTrainingDataset/dataset/lucagplm into the directory: ./dataset/
The training dataset(dataset/lucagplm/v2.0/train/) whose file names start with '2023112418163521' are gene data(DNA + RNA), and those that start with '2023112314061479' are protein data.
The validation dataset(dataset/lucagplm/v2.0/dev/) whose file names start with '2023112418224620' are gene data(DNA + RNA), and those that start with '2023112314080544' are protein data.
The testing dataset(dataset/lucagplm/v2.0/test/) whose file names start with '2023112418231445' are gene data(DNA + RNA), and those that start with '2023112314083364' are protein data.
Notice
If you want to train individual nucleic acid or protein LucaOne(LucaOne-Gene or LucaOne-Prot), please separate the datasets as described above.
Training scripts are under the directory src/training, including 4 shell scripts:
run_multi_v2.0.sh: nucleic acid(DNA+RNA) and protein mixed training with 10 pre-training tasks.
run_multi_mask_v2.0.sh: nucleic acid(DNA+RNA) and protein mixed training with only 2 mask pre-training tasks.
run_multi_v2.0_gene.sh: individual nucleic acid training with 3 pre-training tasks.
run_multi_v2.0_prot.sh: individual protein training with 7 pre-training tasks.
FTP:
Pre-training data, code, and trained checkpoint of LucaOne, embedding inference code, downstream validation tasks data & code, and other materials are available: FTP.
Details:
The LucaOne's model code is available at: LucaOne Github or LucaOne.
The trained-checkpoint files are available at: TrainedCheckPoint.
LucaOne's representational inference code is available at: LucaOneApp Github or LucaOneApp.
The project of 8 downstream tasks is available at: LucaOneTasks Github or LucaOneTasks.
The pre-training dataset of LucaOne is opened at: PreTrainingDataset.
The datasets of downstream tasks are available at: DownstreamTasksDataset .
Other supplementary materials are available at: Others .
Yong He, Zhaorong Li, Pan Fang, Yongtao Shan, Yanhong Wei, Yuan-Fei Pan
@article {LucaOne,
author = {Yong He and Pan Fang and Yongtao Shan and Yuanfei Pan and Yanhong Wei and Yichang Chen and Yihao Chen and Yi Liu and Zhenyu Zeng and Zhan Zhou and Feng Zhu and Edward C. Holmes and Jieping Ye and Jun Li and Yuelong Shu and Mang Shi and Zhaorong Li},
title = {LucaOne: Generalized Biological Foundation Model with Unified Nucleic Acid and Protein Language},
elocation-id = {2024.05.10.592927},
year = {2024},
doi = {10.1101/2024.05.10.592927},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/05/14/2024.05.10.592927},
eprint = {https://www.biorxiv.org/content/early/2024/05/14/2024.05.10.592927.full.pdf},
journal = {bioRxiv}
}