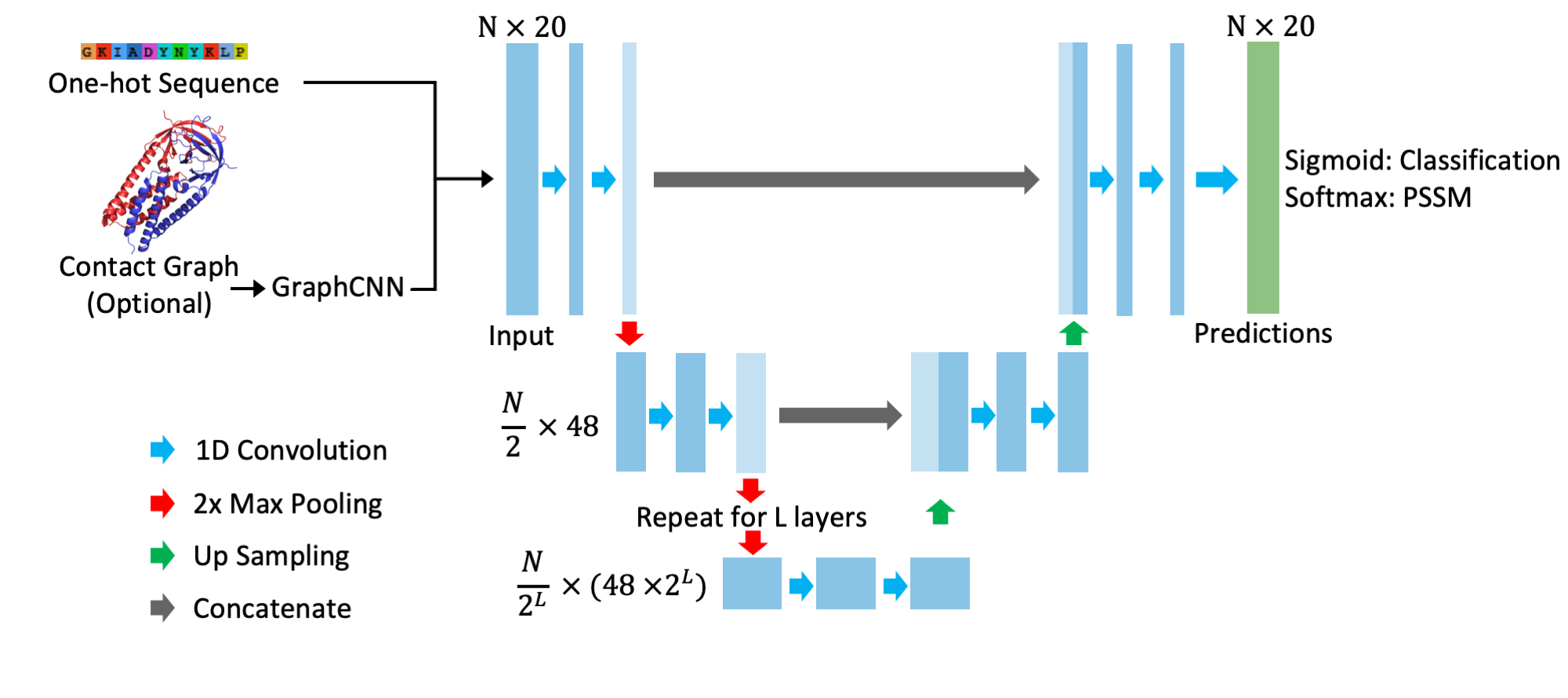
Sequence UNET is a fully convolutional neural network variant effect predictor, able to predict the pathogenicity of protein coding variants and the frequency they occur across large multiple sequence alignments. It uses a U-shaped architecture inspired by the U-NET medical image segmentation network (Ronneberger et al. 201), with an optional Graph CNN section to incorporate information from protein structure:
This repo contains the code that defines the model, various training experiments and analysis of the trained model.
The simplest way to use the model is to clone the repo and add the src directory to your python path.
The important modules and scripts for using the model are:
src/sequence_unet.pytop_model.pygraph_cnn.pymetrics.pypredict.pytrain.py
Saved model weights will be made available soon with a preprint describing the model. In future a python package may be developed to make the process of loading and using the model more straightforward.
The core model requires:
- Python 3
- Tensorflow 2.5+
- Numpy
In addition the training and predictions scripts use:
- Pandas
- Biopython
- ProteinNetPy (Required for training and prediction scripts, not the model itself)
Figure generation and performance analysis is performed in R 4.0, largely using Tidyverse packages.
To initiate an untrained model use the sequence_unet function in sequence_unet.py or one of the top model functions in top_model.py.
These models must be trained manually or have trained weights loaded.
Alternatively a trained model can be loaded directly (when downloads are available), ensuring that the custom_objects from metrics.py is available:
import tensorflow.keras as keras
from metrics import CUSTOM_OBJECTS
model = keras.models.load_model("path/to/model.tf", custom_objects=CUSTOM_OBJECTS)The model functions are currently documented in their docstrings, in future these may be used to generate full documentation as well.
The predict.py script offers two prediction mechanisms, predicting scores for proteins in a fasta or ProteinNet file.
Usage of the script is documented in it's docstring and can be accessed using -h.
The training.py script was used to train the models, based on a saved model and data loading function saved by the make_experiment_dir function in src/utils.py.
Usage of the script is documented in it's docstring and can be accessed using -h.
The training scripts in subdirectories of models give examples of the model training procedures I used to train the various forms of the model.