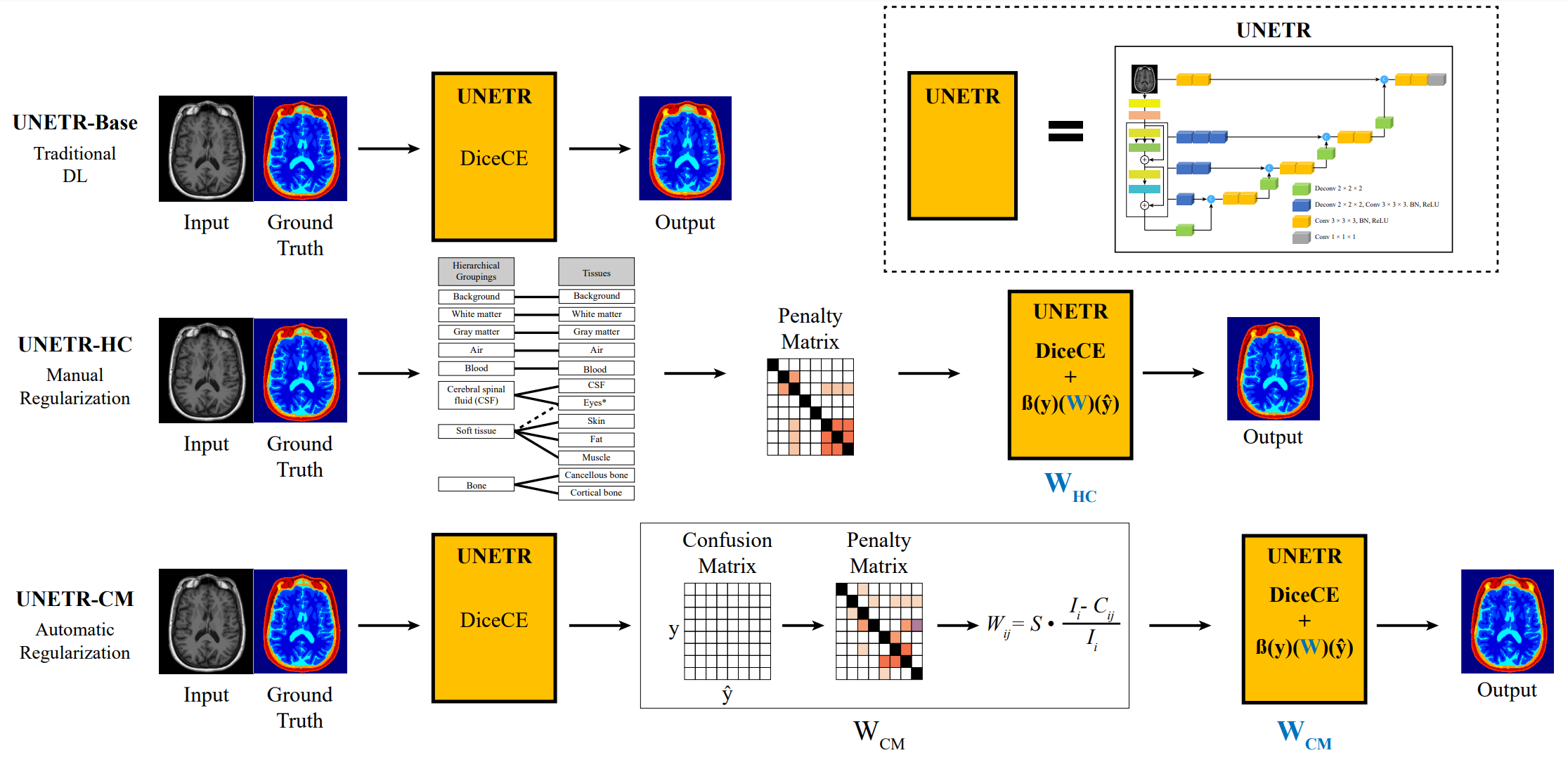
We create and provide open-source code of a model called a domain-aware model calibration method, nicknamed DOMINO that leverages the semantic confusability and hierarchical similarity between class labels. Our experiments demonstrate that our DOMINO-calibrated deep neural networks outperform non-calibrated models and state-of-the-art morphometric methods in head image segmentation. Our results show that our method can consistently achieve better calibration, higher accuracy, and faster inference times than these methods, especially on rarer classes. This performance is attributed to our domain-aware regularization to inform semantic model calibration. These findings show the importance of semantic ties between class labels in building confidence in deep learning models. The framework has the potential to improve the trustworthiness and reliability of generic medical image segmentation models.
This repository provides the official implemantation of training DOMINO as well as the usage the model DOMINO in the following paper:
DOMINO: Domain-aware Model Calibration in Medical Image Segmentation
Skylar E. Stolte1, Kyle Volle2, Aprinda Indahlastari3,4, Alejandro Albizu3,5, Adam J. Woods3,4,5, Kevin Brink6, Matthew Hale2, and Ruogu Fang1,3,7*
1 J. Crayton Pruitt Family Department of Biomedical Engineering, HerbertWertheim College of Engineering, University of Florida (UF), USA
2 Department of Mechanical and Aerospace Engineering, Herbert Wertheim Collegeof Engineering, UF, USA
3 Center for Cognitive Aging and Memory, McKnight Brain Institute, UF, USA
4 Department of Clinical and Health Psychology, College of Public Health andHealth Professions, UF, USA
5 Department of Neuroscience, College of Medicine, UF, USA
6 United States Air Force Research Laboratory, Eglin Air Force Base, Florida, USA
7 Department of Electrical and Computer Engineering, Herbert Wertheim College ofEngineering, UF, USA
International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2022
paper | code | slides | poster | talk
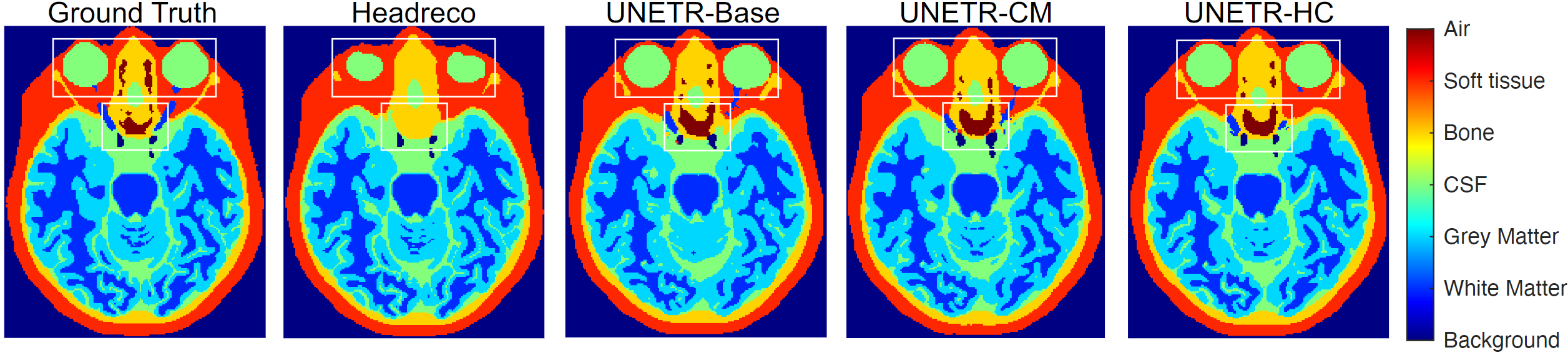
- Our DOMINO methods improve calibration and accuracy in head segmentation problems from T1 MRIs
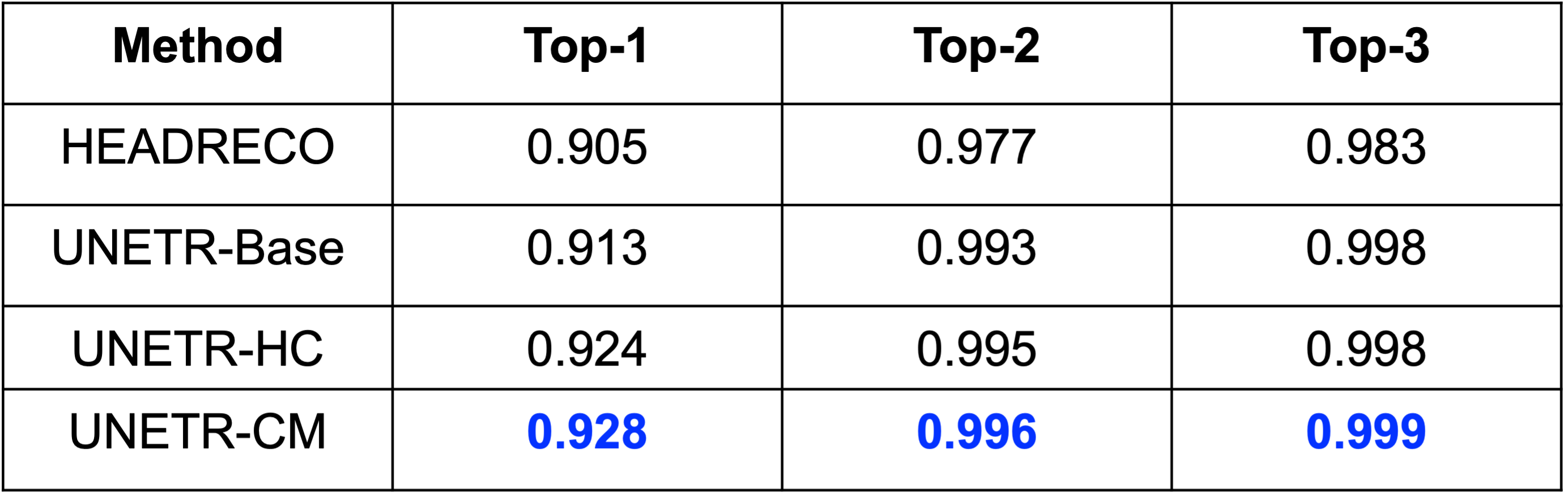
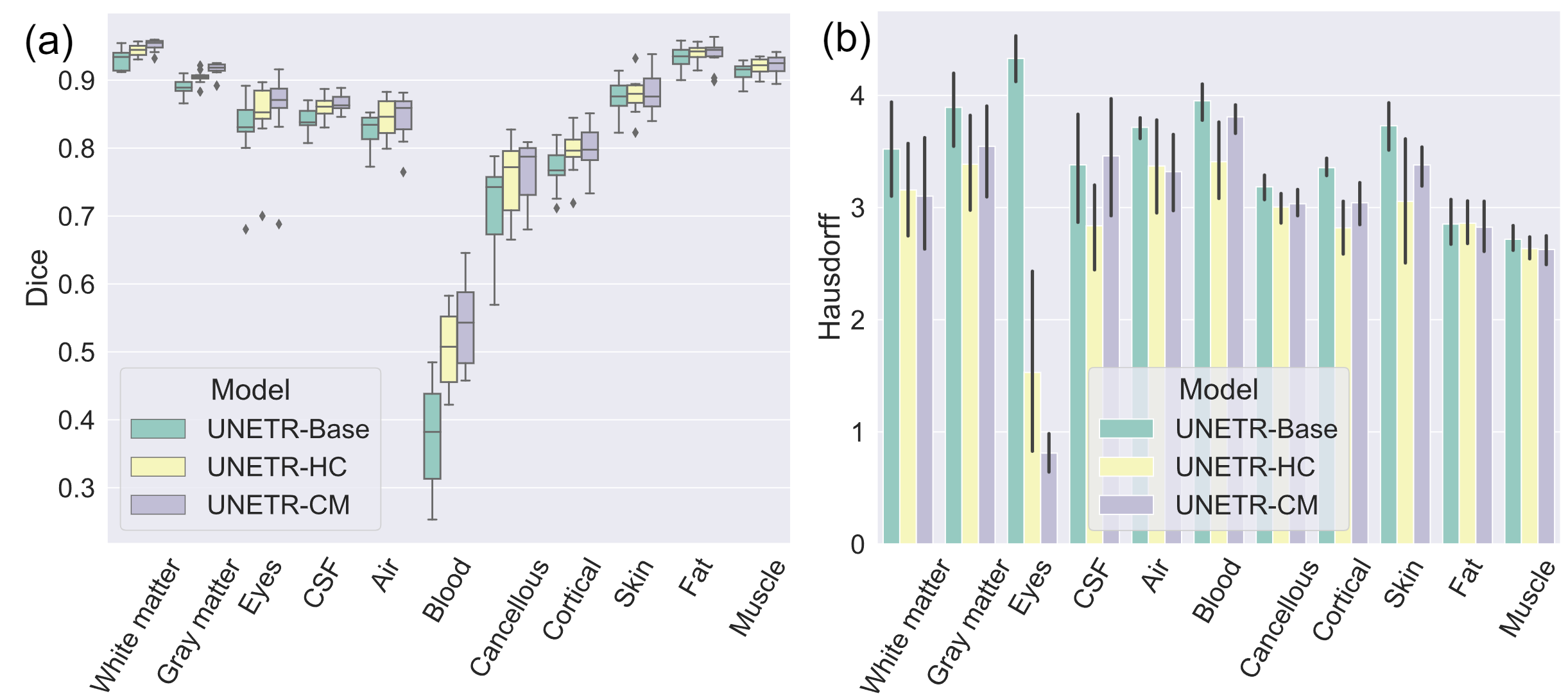
- DOMINO-CM achieves higher Top-1, Top-2, and Top-3 accuracy than DOMINO-HC or an uncalibrated model. This indicates superior regional performance and higher relevance to the non-selected classes (important in calibration).
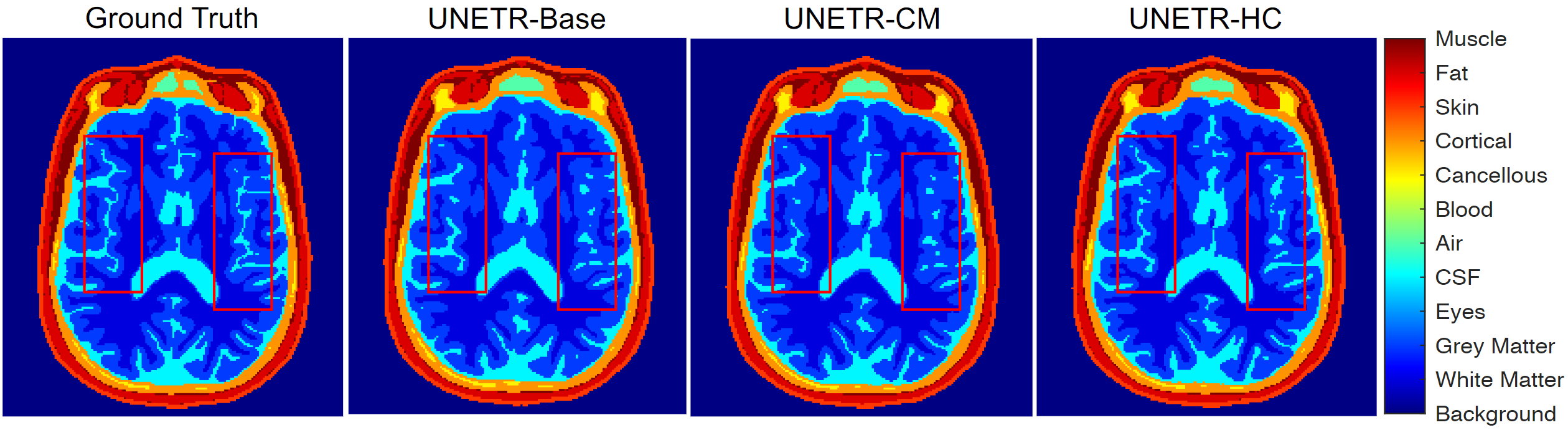
- DOMINO-HC achieves more precise boundary detection when compared to DOMINO-CM or an uncalibrated model. This is important to calibration problems because boundaries are the most uncertain areas in segmentation challenges.
You can find there are two MATLAB codes, you can directly change the directory to your own data. You need to select the DOMINO working folder and add to path before you running these two MATLAB codes.
To run the combineIn case of you are using different version of MATLAB, if you are using MATLAB 2020b, you need to change line 56 to :
image(index) = tissue_cond_updated.Labels(k)
Then you can run the combine_mask.m. The output should be a Data folder with the following structure:
Data ImagesTr sub-TrX_T1.nii sub-TrXX_T1.nii ...
ImagesTs sub-TsX_T1.nii sub-TsXX_T1.nii ...
LabelsTr sub-TrX_seg.nii sub-TrXX_seg.nii ...
LabelsTs sub-TsX_seg.nii sub-TsX_seg.nii ...
Maneuver to the /your_data/Data/. Run make_datalist_json.m
After this code is done, you may exit MATLAB and open the terminal to run the other codes.
The DOMINO code uses the MONAI, an open-source foundation. We provide a .sh script to help you to build your own container for running your code.
Run the following code in the terminal, you need to change the line after --sandbox to your desired writable directory and change the line after --nv to your own directory.
sbatch building_container_v08.sh
The output should be a folder named monaicore08 under your desired directory.
Once the data and the container are ready, you can train the model by using the following command:
sbatch train.sh
Before you training the model, you need to make sure change the following directory:
- change the first singularity exec -nv to the directory includes monaicore08, for example: /user/DOMINO/monaicore08
- change the line after --bind to the directory includes monaicore08
- change the data_dir to your data directory
- change the model name to your desired model name You can also specify the max iteration number for training. For the iterations = 100, the training progress might take about one hours, and for the iterations = 25,000, the training progress might take about 24 hours.
The test progress is very similar to the training progress. You need to change all paths and make sure the model_save_name matches your model name in runMONAI.sh. Then running the runMONAI_test.sh with the following command:
sbatch test.sh
The outputs for each test subject is saved as a mat file.
You can also use the pre-trained models we provide for testing, please fill out the following request form before accessing to DOMINO models. Download pre-trained models here
You can alternatively find the link to our reproducible capsule for our code on Code Ocean. The link to this capsule is as follows: https://codeocean.com/capsule/6022409/tree/v2.
If you use this code, please cite our papers:
@InProceedings{stolte2022DOMINO,
author="Stolte, Skylar E. and Volle, Kyle and Indahlastari, Aprinda and Albizu, Alejandro and Woods, Adam J. and Brink, Kevin and Hale, Matthew and Fang, Ruogu",
title="DOMINO: Domain-aware Model Calibration in Medical Image Segmentation",
booktitle="Medical Image Computing and Computer Assisted Intervention (MICCAI) 2022",
year="2022",
url="https://arxiv.org/abs/2209.06077"
}
This work was supported by the National Institutes ofHealth/National Institute on Aging (NIA RF1AG071469, NIA R01AG054077),the National Science Foundation (1908299), and the NSF-AFRL INTERN Sup-plement (2130885).
We acknowledge NVIDIA AI Technology Center (NVAITC)for their suggestions. We also thank Jiaqing Zhang for formatting assistance.
We employ UNETR as our base model from: https://github.com/Project-MONAI/research-contributions/tree/main/UNETR
Any discussion, suggestions and questions please contact: Skylar Stolte, Dr. Ruogu Fang.
Smart Medical Informatics Learning & Evaluation Laboratory, Dept. of Biomedical Engineering, University of Florida