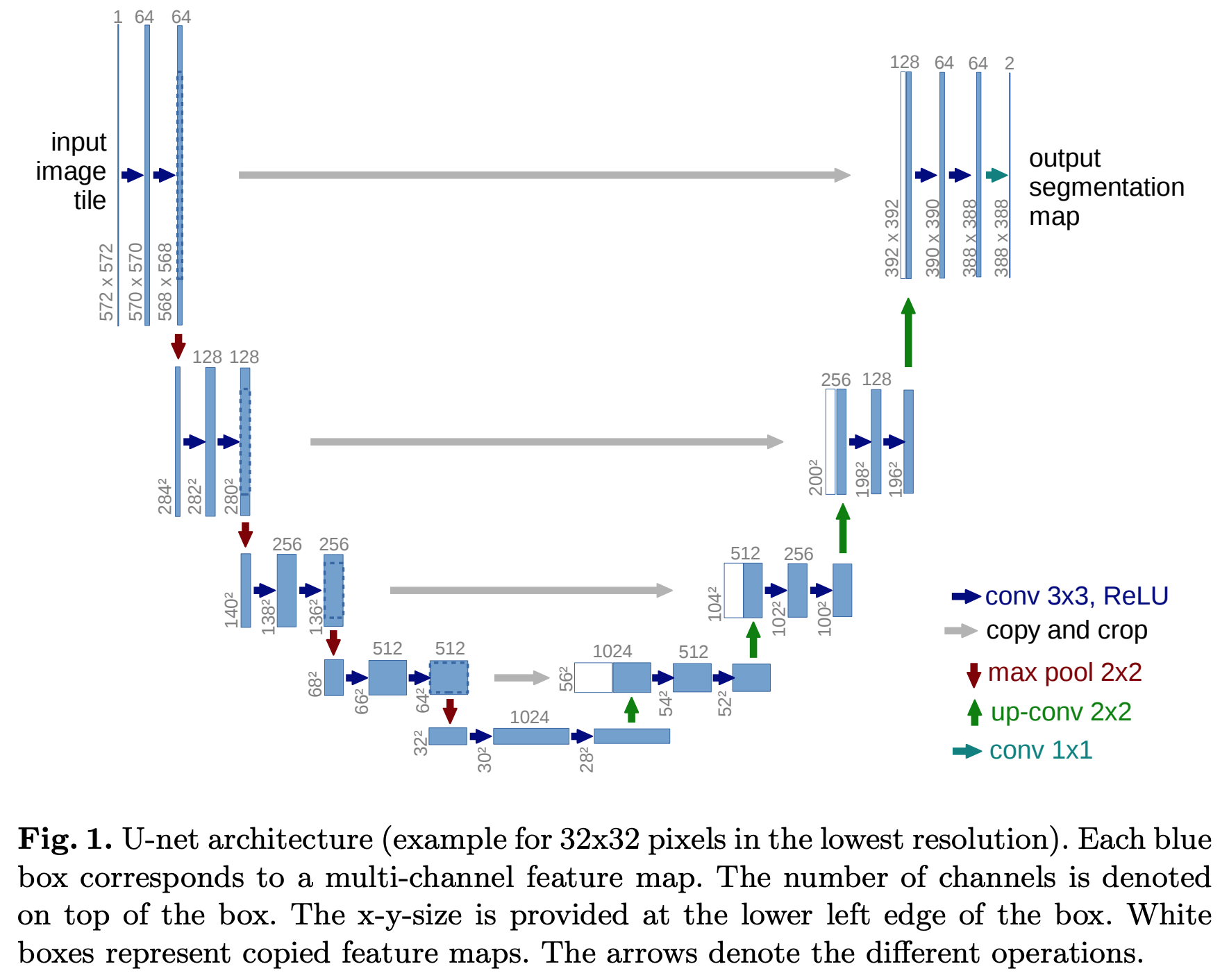
U-Net: Convolutional Networks for Biomedical Image Segmentation
There is large consent that successful training of deep networks requires many thousand annotated training samples. In this paper, we present a network and training strategy that relies on the strong use of data augmentation to use the available annotated samples more efficiently. The architecture consists of a contracting path to capture context and a symmetric expanding path that enables precise localization. We show that such a network can be trained end-to-end from very few images and outperforms the prior best method (a sliding-window convolutional network) on the ISBI challenge for segmentation of neuronal structures in electron microscopic stacks. Using the same network trained on transmitted light microscopy images (phase contrast and DIC) we won the ISBI cell tracking challenge 2015 in these categories by a large margin. Moreover, the network is fast. Segmentation of a 512x512 image takes less than a second on a recent GPU. The full implementation (based on Caffe) and the trained networks are available at this http URL.
- Before running Train phase, execute run_make_database.py to generate dataset.
$ python main.py --mode train \
--scope [scope name] \
--name_data [data name] \
--dir_data [data directory] \
--dir_log [log directory] \
--dir_checkpoint [checkpoint directory]
--gpu_ids [gpu id; '-1': no gpu, '0, 1, ..., N-1': gpus]
$ python main.py --mode train \
--scope unet \
--name_data em \
--dir_data ./datasets \
--dir_log ./log \
--dir_checkpoint ./checkpoint
--gpu_ids 0
- Set [scope name] uniquely.
- To understand hierarchy of directories based on their arguments, see directories structure below.
- Hyperparameters were written to arg.txt under the [log directory].
$ python main.py --mode test \
--scope [scope name] \
--name_data [data name] \
--dir_data [data directory] \
--dir_log [log directory] \
--dir_checkpoint [checkpoint directory] \
--dir_result [result directory]
--gpu_ids [gpu id; '-1': no gpu, '0, 1, ..., N-1': gpus]
$ python main.py --mode test \
--scope unet \
--name_data em \
--dir_data ./datasets \
--dir_log ./log \
--dir_checkpoint ./checkpoints \
--dir_result ./results
--gpu_ids 0
- To test using trained network, set [scope name] defined in the train phase.
- Generated images are saved in the images subfolder along with [result directory] folder.
- index.html is also generated to display the generated images.
$ tensorboard --logdir [log directory]/[scope name]/[data name] \
--port [(optional) 4 digit port number]
$ tensorboard --logdir ./log/unet/em \
--port 6006
After the above comment executes, go http://localhost:6006
- You can change [(optional) 4 digit port number].
- Default 4 digit port number is 6006.
1st row: input; serial section Transmission Electron Microscopy (ssTEM)
2nd row: label; sementation map
3rd row: output; predection map by unet
- The results were generated by a network trained with em dataset during 300 epochs.
- After running the Test phase, execute display_result.py to display the figure.
pytorch-UNET
+---[dir_checkpoint]
| \---[scope]
| \---[name_data]
| +---model_epoch00000.pth
| | ...
| \---model_epoch12345.pth
+---[dir_data]
| \---[name_data]
| +---test
| | +---00000.npy
| | | ...
| | \---12345.npy
| +---train
| | +---00000.npy
| | | ...
| | \---12345.npy
| \---val
| +---00000.npy
| | ...
| \---12345.npy
+---[dir_log]
| \---[scope]
| \---[name_data]
| +---arg.txt
| \---events.out.tfevents
\---[dir_result]
\---[scope]
\---[name_data]
+---images
| +---00000-input.png
| +---00000-label.png
| +---00000-output.png
| | ...
| +---12345-input.png
| +---12345-label.png
| +---12345-output.png
\---index.html
pytorch-UNET
+---checkpoints
| \---unet
| \---em
| +---model_epoch0000.pth
| | ...
| \---model_epoch0300.pth
+---datasets
| \---em
| +---test
| | +---0.npy
| | | ...
| | \---4.npy
| +---train
| | +---0.npy
| | | ...
| | \---29.npy
| \---val
| +---0.npy
| | ...
| \---4.npy
+---log
| \---unet
| \---em
| +---arg.txt
| \---events.out.tfevents
\---results
\---unet
\---em
+---images
| +---0000-input.png
| +---0000-label.png
| +---0000-output.png
| | ...
| +---0004-input.png
| +---0004-label.png
| +---0004-output.png
\---index.html
- Above directory is created by setting arguments when main.py is executed.