SynthSeg
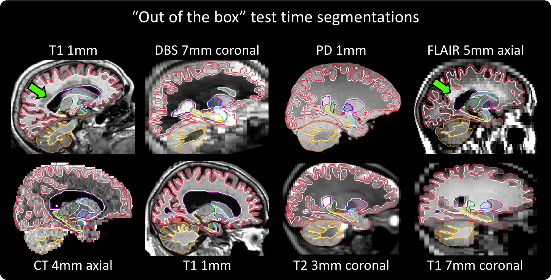
In this repository, we present SynthSeg, the first convolutional neural network for segmentation of brain scans of any contrast and resolution that works out-of-the-box, without retraining or fine-tuning. SynthSeg relies on a single model, which is robust to:
- a wide array of subject populations: from young and healthy to ageing and diseased subjects,
- white matter lesions,
- scans with or without preprocessing, including bias field corruption, skull stripping, intensity normalisation, template registration, etc.
Here, we distribute the open-source model along with the corresponding code to enable researchers to run SynthSeg on
their own data. We emphasise that predictions are given at 1mm isotropic resolution (regardless of the resolution of the
input images), and can be obtained either by running on the GPU (6s per scan) or on the CPU (1min).
New features and updates
04/10/2022: SynthSeg is available with Matlab! ⭐
We are delighted, as much as surprised, that Matlab 2022b (and onwards) now includes SynthSeg in its Medical Image
Toolbox. They have a fully documented example on how to use it
here.
But, to simplify things, we wrote our own Matlab wrapper, which you can call in one single line. Just download this zip
file by clicking
here,
uncompress it, open Matlab, and type help SynthSeg for instructions.
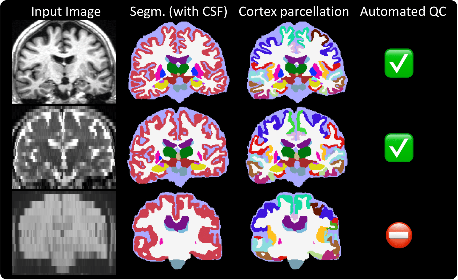
29/06/2022: SynthSeg 2.0 is out ! ✌️
In addition to whole-brain segmentation, it now also performs Cortical parcellation, automated QC, and intracranial
volume (ICV) estimation (see figure below).
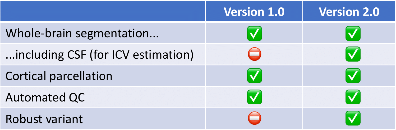
For practical reasons, most of these features are compatible with SynthSeg 1.0. Here is a table with a summary of the
functionalities supported by each version.
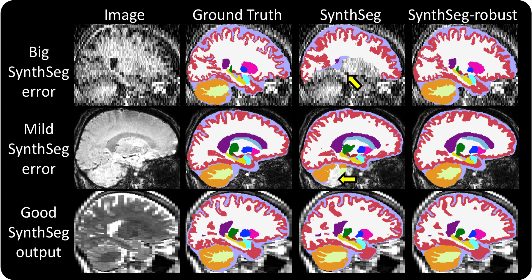
01/03/2022: Robust version 🔨
SynthSeg sometimes falters on scans with low signal-to-noise ratio, or with very low tissue contrast. For this reason,
we developed a new model for increased robustness, named "SynthSeg-robust". You can use this mode when SynthSeg gives
results like in the figure below:
29/10/2021: SynthSeg is now available on the dev version of
FreeSurfer !! 🎉
See here on how to use it.
Try it in one command !
Once all the python packages are installed (see below), you can simply test SynthSeg on your own data with:
python ./scripts/commands/SynthSeg_predict.py --i <input> --o <output> [--parc --robust --vol <vol> --qc <qc> --post <post> --resample <resample>]
where:
<input>path to a scan to segment, or to a folder. This can also be the path to a text file, where each line is the path of an image to segment.<output>path where the output segmentations will be saved. This must be the same type as<input>(i.e., the path to a file, a folder, or a text file where each line is the path to an output segmentation).--parc(optional) to perform cortical parcellation in addition to whole-brain segmentation.--robust(optional) to use the variant for increased robustness (e.g., when analysing clinical data with large space spacing). This can be slower than the other model.<vol>(optional) path to a CSV file where the volumes of all segmented regions will be saved for all scans (e.g. /path/to/volumes.csv). If<input>is a text file, so must be<vol>, for which each line is the path to a different CSV file corresponding to one subject only.<qc>(optional) path to a CSV file where QC scores will be saved. The same formatting requirements apply as for<vol>.<post>(optional) path where the posteriors, given as soft probability maps, will be saved (same formatting requirements as for<output>).<resample>(optional) SynthSeg segmentations are always given at 1mm isotropic resolution. Hence, images are always resampled internally to this resolution (except if they are already at 1mm resolution). Use this flag to save the resampled images (same formatting requirements as for<output>).
Additional optional flags are also available:
--cpu: (optional) to enforce the code to run on the CPU, even if a GPU is available.--threads: (optional) number of threads to be used by Tensorflow (default uses one core). Increase it to decrease the runtime when using the CPU version.--crop: (optional) to crop the input images to a given shape before segmentation. This must be divisible by 32. Images are cropped around their centre, and their segmentations are given at the original size. It can be given as a single (i.e.,--crop 160), or several integers (i.e,--crop 160 128 192, ordered in RAS coordinates). By default the whole image is processed. Use this flag for faster analysis or to fit in your GPU.--fast: (optional) to disable some operations for faster prediction (twice as fast, but slightly less accurate). This doesn't apply when the --robust flag is used.--v1: (optional) to run the first version of SynthSeg (SynthSeg 1.0, updated 29/06/2022).
IMPORTANT: SynthSeg always give results at 1mm isotropic resolution, regardless of the input. However, this can
cause some viewers to not correctly overlay segmentations on their corresponding images. In this case, you can use the
--resample flag to obtain a resampled image that lives in the same space as the segmentation, such that they can be
visualised together with any viewer.
The complete list of segmented structures is available in labels table.txt along with their corresponding values. This table also details the order in which the posteriors maps are sorted.
Installation
-
Clone this repository
-
Create a virtual environment, and install all the required packages. We initially gave all the required packages to make it work with Python 3.6 (see requirements_3.6), but we now also give the list for Python 3.8 (see requirements_3.8). Therefore, the choice is yours, but in each case, please stick to the exact versions of the corresponding packages. If you wish to run SynthSeg on the GPU, or to train your own model, you will also need the usual deep learning libraries Cuda (10.0), and CUDNN (7.0).
-
Go to this link UCL dropbox, and download the missing models. Then simply copy them to models.
That's it ! You're now ready to use this tool ! 🎉
How does it work ?
In short, we train a network with synthetic images sampled on the fly from a generative model based on the forward model of Bayesian segmentation. Crucially, we adopt a domain randomisation strategy where we fully randomise the generation parameters which are drawn at each minibatch from uninformative uniform priors. By exposing the network to extremely variable input data, we force it to learn domain-agnostic features. As a result, SynthSeg is able to readily segment real scans of any target domain, without retraining or fine-tuning.
The following figure first illustrates the workflow of a training iteration, and then provides an overview of the
different steps of the generative model:
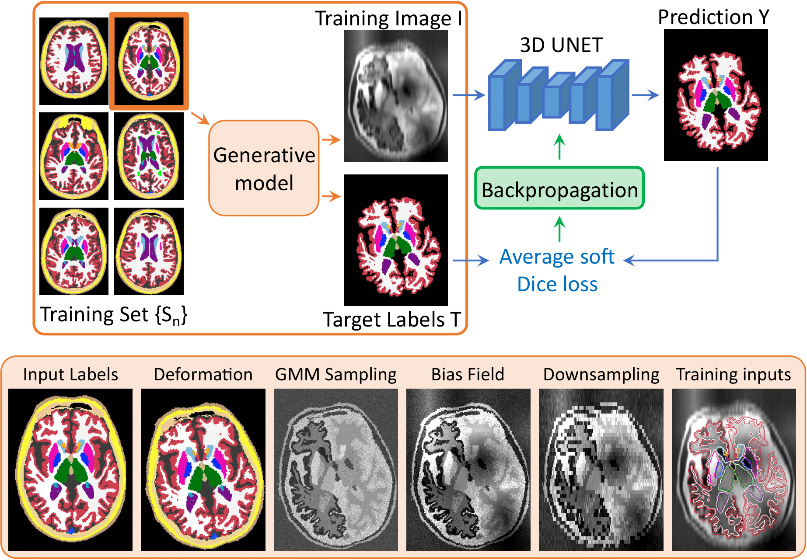
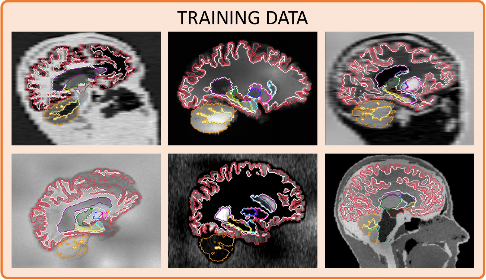
Finally we show additional examples of the synthesised images along with an overlay of their target segmentations:
If you are interested to learn more about SynthSeg, you can read the associated publication (see below), and watch this
presentation, which was given at MIDL 2020 for a related article on a preliminary version of SynthSeg (robustness to
MR contrast but not resolution).

Train your own model
This repository contains all the code and data necessary to train, validate, and test your own network. Importantly, the proposed method only requires a set of anatomical segmentations to be trained (no images), which we include in data. While the provided functions are thoroughly documented, we highly recommend to start with the following tutorials:
-
1-generation_visualisation: This very simple script shows examples of the synthetic images used to train SynthSeg.
-
2-generation_explained: This second script describes all the parameters used to control the generative model. We advise you to thoroughly follow this tutorial, as it is essential to understand how the synthetic data is formed before you start training your own models.
-
3-training: This scripts re-uses the parameters explained in the previous tutorial and focuses on the learning/architecture parameters. The script here is the very one we used to train SynthSeg !
-
4-prediction: This scripts shows how to make predictions, once the network has been trained.
-
5-generation_advanced: Here we detail more advanced generation options, in the case of training a version of SynthSeg that is specific to a given contrast and/or resolution (although these types of variants were shown to be outperformed by the SynthSeg model trained in the 3rd tutorial).
-
6-intensity_estimation: This script shows how to estimate the Gaussian priors of the GMM when training a contrast-specific version of SynthSeg.
-
7-synthseg+: Finally, we show how the robust version of SynthSeg was trained.
These tutorials cover a lot of materials and will enable you to train your own SynthSeg model. Moreover, even more detailed information is provided in the docstrings of all functions, so don't hesitate to have a look at these !
Content
-
SynthSeg: this is the main folder containing the generative model and training function:
-
labels_to_image_model.py: contains the generative model for MRI scans.
-
brain_generator.py: contains the class
BrainGenerator, which is a wrapper aroundlabels_to_image_model. New images can simply be generated by instantiating an object of this class, and call the methodgenerate_image(). -
training.py: contains code to train the segmentation network (with explanations for all training parameters). This function also shows how to integrate the generative model in a training setting.
-
predict.py: prediction and testing.
-
validate.py: includes code for validation (which has to be done offline on real images).
-
-
models: this is where you will find the trained model for SynthSeg.
-
data: this folder contains some examples of brain label maps if you wish to train your own SynthSeg model.
-
script: contains tutorials as well as scripts to launch trainings and testings from a terminal.
-
ext: includes external packages, especially the lab2im package, and a modified version of neuron.
Citation/Contact
This code is under Apache 2.0 licensing. If you use it, please cite the following paper:
SynthSeg: Domain Randomisation for Segmentation of Brain MRI Scans of any Contrast and Resolution
B. Billot, D.N. Greve, O. Puonti, A. Thielscher, K. Van Leemput, B. Fischl, A.V. Dalca, J.E. Iglesias
[arxiv | bibtex]
Instead, if you use the cortical parcellation, automated QC, or robust version, please cite the following paper:
Robust Segmentation of Brain MRI in the Wild with Hierarchical CNNs and no Retraining
B. Billot, M. Colin, S.E. Arnold, S. Das, J.E. Iglesias
MICCAI 2022
[arxiv | bibtex]
If you have any question regarding the usage of this code, or any suggestions to improve it, you can contact us at:
benjamin.billot.18@ucl.ac.uk