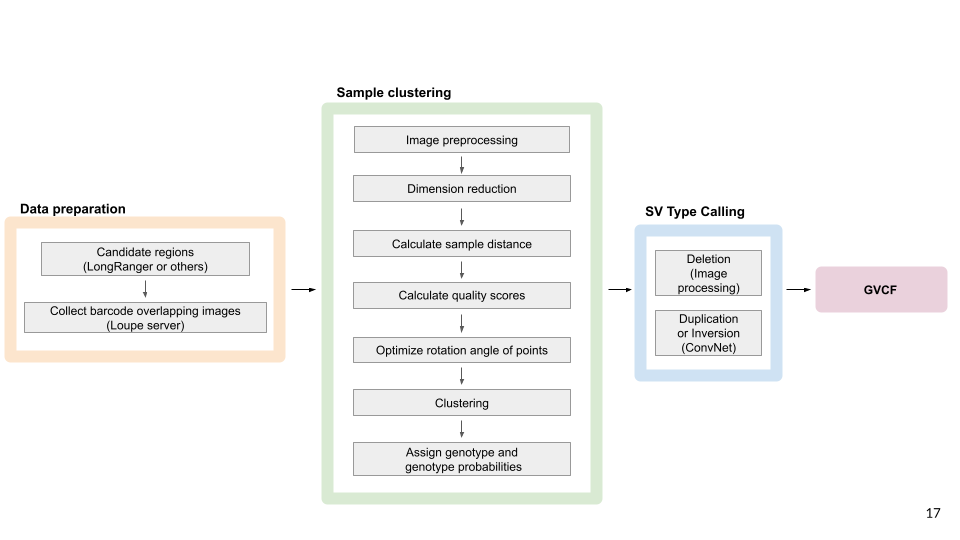
SVJAM jointly detects and genotypes large structural variants (SVs) from linked-read whole genome sequence data.
SVJAM was developed on python 3.8
Run the following command to clone the repository
git clone https://github.com/hakangunturkun/SVJAM
Then use the following command to install all the required libraries
cd SVJAM
pip install -r requirements.txt
You'll also need a copy of the reference genome that matches your LongRanger analysis and place that in the RefGenome folder (file needs to be unzipped, with the extension name .fa)
We provide an example image set for testing in chr1/images/ folder. Image files are organized in folders named as: chr1_93011035-93221780_chr1_93011035-93221780 The output will be located in the chr1/output/ folder.
You need to download the mouse reference genome from UCSC, uncompress it, and put it in the RefGenome folder.
To do a test run using this data set, run the following command from the SVJAM installation directory
python SVJAM.py chr1
- Install the loupe browser from 10X Genomics.
- Generate *.loupe files from linked-read WGS by using longranger.
- Open and organize image_download/open_loupe.sh then run it.
- Download images manually or use image_download/chrome_multi_save_matrixView_pngs.py for automatically download that uses Selenium library.
Gunturkun, M.H., Villani, F., Colonna, V., Ashbrook, D., Williams, R.W. and Chen, H. (2021). SVJAM: Joint analysis of structural variants usinglinked read sequencing data. bioRxiv. doi:10.1101/2021.11.02.467006