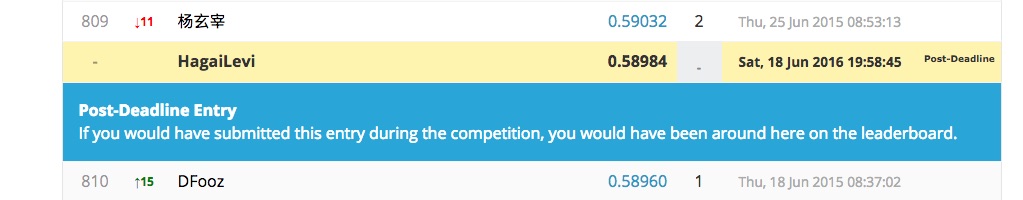
We have tried 2 models, one is simplistic, fast and less accurate, and the other is really compute intensive, slow and provides better prediction.
We will build a model and try to predict the median_relevance. We will clean the text from punctuation html tags etc, then for each
row, we will rank its relevance by checking how many of the words that appear in the query, also appear in the description and title of the product,
and this will be our smart feature.
This is the more simplistic model.
We clean the text, and check how many of the words that appear in the query also appear in the description and title
library(readr)
library(tm)train <- read_csv("data/train.csv")
test <- read_csv("data/test.csv")We define a function to clean up our text
cleanText <- function(x) {
# remove html tags
x = gsub("<.*?>", "", x)
# lower case
x = tolower(x)
# remove numbers
x = gsub("[[:digit:]]", "", x)
# remove punctuation symbols
x = gsub("[[:punct:]]", "", x)
# remove numbers
x = removeNumbers(x)
# remove stop words
x = removeWords(x, stopwords("en"))
# remove extra white spaces
x = gsub("[ \t]{2,}", " ", x)
x = gsub("^\\s+|\\s+$", "", x)
return (x)
}We define functions that calculate the correlation between 2 strings, by checking how many of the words that appear in the first string, also appear in the second.
# This function calculates the percentage of words that appear in the query and in the title
calcRelevance <- function(query, title, description) {
matching <- 0
for (word in strsplit(query, " ")[[1]]) {
if (grepl(word, title)) {
matching <- matching+1;
}
}
return(matching / length(strsplit(query, " ")[[1]]))
}
# an abstraction for calcRelevance for query and product title
calcRowRelevance <- function(row) {
calcRelevance(row[['query']],row[['product_title']],row[['product_description']])
}
# an abstraction for calcRelevance for query and product description
calcRowRelevance2 <- function(row) {
calcRelevance(row[['query']],row[['product_description']],"")
}Clean up and preprocess the data
# convert median_relevance to a factor as it is not continuos
train$median_relevance <- factor(train$median_relevance)
#Preprocess the train data
train$query <- cleanText(train$query)
train$product_title <- cleanText(train$product_title)
train$product_description <- cleanText(train$product_description)
train$relevance <- apply(train, 1, FUN=calcRowRelevance)
train$relevance2 <- apply(train, 1, FUN=calcRowRelevance2)
#Preprocess the test data as well
test$query <- cleanText(test$query)
test$product_title <- cleanText(test$product_title)
test$product_description <- cleanText(test$product_description)
test$relevance <- apply(test, 1, FUN=calcRowRelevance)
test$relevance2 <- apply(test, 1, FUN=calcRowRelevance2)
# In order to avoid tackling test categories that are unfamiliar to the trained model, we make sure that the nominal
# attribute is set according to categories in both the train and test sets.
levels(train$query) <- union(levels(train$query), levels(test$query))
levels(train$product_title) <- union(levels(train$product_title), levels(test$product_title))
levels(train$product_description) <- union(levels(train$product_description), levels(test$product_description))
levels(test$query) <- union(levels(train$query), levels(test$query))
levels(test$product_title) <- union(levels(train$product_title), levels(test$product_title))
levels(test$product_description) <- union(levels(train$product_description), levels(test$product_description))Build the model
inTraining <- sample(1:nrow(train), .75*nrow(train))
training <- train[ inTraining,]
testing <- train[-inTraining,]
# Random forest
library(randomForest)## randomForest 4.6-12
## Type rfNews() to see new features/changes/bug fixes.
model1 <- randomForest(median_relevance ~ relevance, data=training, ntree=3)
library(party)## Loading required package: grid
## Loading required package: mvtnorm
## Loading required package: modeltools
## Loading required package: stats4
## Loading required package: strucchange
## Loading required package: zoo
##
## Attaching package: 'zoo'
## The following objects are masked from 'package:base':
##
## as.Date, as.Date.numeric
## Loading required package: sandwich
library(caret)## Loading required package: lattice
## Loading required package: ggplot2
##
## Attaching package: 'ggplot2'
## The following object is masked from 'package:randomForest':
##
## margin
## The following object is masked from 'package:NLP':
##
## annotate
model2 <- train(median_relevance ~ relevance, data = training,
method = "rpart",
trControl = trainControl(classProbs = F))## Loading required package: rpart
results1 <- predict(model1, newdata = testing)
results2 <- predict(model2, newdata = testing)
library(Metrics)
ScoreQuadraticWeightedKappa(testing$median_relevance, results1, 1, 4)## [1] 0.01712326
ScoreQuadraticWeightedKappa(testing$median_relevance, results2, 1, 4)## [1] 0.3869481
results1 <- predict(model1, newdata = test)
Newsubmission = data.frame(id=test$id, prediction = results1)
write.csv(Newsubmission,"model1.csv",row.names=F)
results2 <- predict(model2, newdata = test)
Newsubmission = data.frame(id=test$id, prediction = results2)
write.csv(Newsubmission,"model2.csv",row.names=F) That is our first model, it provided results of about 0.35
Our other model is super complicated (computation wise), and takes about 3 days to run on an ec2 instance with 8 cores and 60 GB ram.
We ran it as an .R file, and not as .Rmd file, and we didn't want to run it again (costs a lot of money), so the code is attached, but we
didn't actually ran it with the markdown file, but only once with the R file.
In order to simulate a run of the model, just change eval=FALSE to eval=TRUE in the following blocks, and un-comment the lines
train <- train[sample(nrow(train), 100), ] and test <- test[sample(nrow(test), 200), ]. This way, you will use only small part of the data
to build the model (100 rows from the training set and 200 from the test) and it will be way faster (but less accurate). This is how we have developed this model
Initialization:
library(readr)
train <- read_csv("data/train.csv")
test <- read_csv("data/test.csv")
set.seed(0)
# train <- train[sample(nrow(train), 100), ]
N.train <- dim(train)[[1]]
# test <- test[sample(nrow(test), 200), ]
N.test <- dim(test)[[1]]
print("finished reading the data")Pre processing the data
# put all the titles (test and train) in single list
doc.list <- c(as.list(train$product_title),as.list(test$product_title))
# put all the queries (test and train) in single list
query.list <- c(as.list(train$query),as.list(test$query))
N.docs <- length(doc.list)
print("Got total docs:")
print(N.docs)Creating the corpus and clean it:
# Create a corpus from all the text, and clean it from punctuation and numbers + stem it
library(tm)
my.docs <- VectorSource(c(doc.list, query.list))
my.docs$Names <- c(names(doc.list), seq(1,length(query.list)))
my.corpus <- Corpus(my.docs)
print("finished creating corpus")
my.corpus <- tm_map(my.corpus, removePunctuation)
my.corpus <- tm_map(my.corpus, stemDocument)
my.corpus <- tm_map(my.corpus, removeNumbers)
print("stem + remove numbers")
my.corpus <- tm_map(my.corpus, content_transformer(tolower))
my.corpus <- tm_map(my.corpus, stripWhitespace)
print("finished cleaning")Build a term document matrix
# Build a TDM
term.doc.matrix.stm <- TermDocumentMatrix(my.corpus, control = list(minWordLength = 1))
print("created TermDocumentMatrix")
term.doc.matrix <- as.matrix(term.doc.matrix.stm)
calc_weights <- function(tf.vec, df) {
# Computes tfidf weights from a term frequency vector and a document
# frequency scalar
weight = rep(0, length(tf.vec))
weight[tf.vec > 0] = (1 + log2(tf.vec[tf.vec > 0])) * log2(N.docs/df)
weight
}
calc_term_vec_weight <- function(tfidf.row) {
# Calculate the weight for the terms in the vector
term.df <- sum(tfidf.row[1:N.docs] > 0)
tf.idf.vec <- calc_weights(tfidf.row, term.df)
return(tf.idf.vec)
}
# build a matrix
tfidf.matrix <- t(apply(term.doc.matrix, c(1), FUN = calc_term_vec_weight))
colnames(tfidf.matrix) <- colnames(term.doc.matrix)
tfidf.matrix <- scale(tfidf.matrix, center = FALSE, scale = sqrt(colSums(tfidf.matrix^2)))
print("finished to build the matrix")
# Keep an original for all the modifications in each iteration
original.tfidf.matrix <- tfidf.matrix
values = list()Calculate our rank for each row. It is based on the relatedness between the search query and the product title.
We couldn't use the product description because it takes 100s of GB of ram to build a DTM based on the description.
print("Starting to iterate")
# calculate the relevance for each document
for (index in seq(1,N.docs)) {
if(index %% 100 == 0) {
print(index)
}
query.vector <- original.tfidf.matrix[, (N.docs + index)]
tfidf.matrix <- tfidf.matrix[, 1:N.docs]
# matrix multiplication
doc.scores <- t(query.vector) %*% tfidf.matrix
value <- doc.scores[[index]]
values <- c(values,value)
}Clean up the results:
# replace Nan with 0
values <- rapply( values, f=function(x) ifelse(is.nan(x),0,x), how="replace" )
train2 <- train
# Save the new data to the original datasets
for(i in seq(1,N.train)){
train2[i,'xxx'] <- values[[i]]
}
for(i in seq(1,N.test)){
test[i,'xxx'] <- values[[N.train + i]]
}Train models, evaluate them, and output the predictions
inTraining <- sample(1:nrow(train2), .75*nrow(train2))
training <- train2[ inTraining,]
testing <- train2[-inTraining,]
# Random forest
library(randomForest)
library(Metrics)
# Build 2 models
model1 <- randomForest(median_relevance ~ xxx, data=training, ntree=3)
results1 <- predict(model1, newdata = testing)
print("results for 1st model:")
print(ScoreQuadraticWeightedKappa(testing$median_relevance, results1, 1, 4))
results1 <- predict(model1, newdata = test)
Newsubmission = data.frame(id=test$id, prediction = results1)
write.csv(Newsubmission,"model1.csv",row.names=F)
model2 <- randomForest(median_relevance ~ xxx, data=training, ntree=5)
results2 <- predict(model2, newdata = testing)
print("results for 2nd model:")
print(ScoreQuadraticWeightedKappa(testing$median_relevance, results2, 1, 4))
results2 <- predict(model2, newdata = test)
Newsubmission = data.frame(id=test$id, prediction = results2)
write.csv(Newsubmission,"model2.csv",row.names=F) In the end, we have used the second model with ntree=3.
This model got us results of about 0.558.
Its results are in final_submission.csv, and those are the results that we used in kaggle.