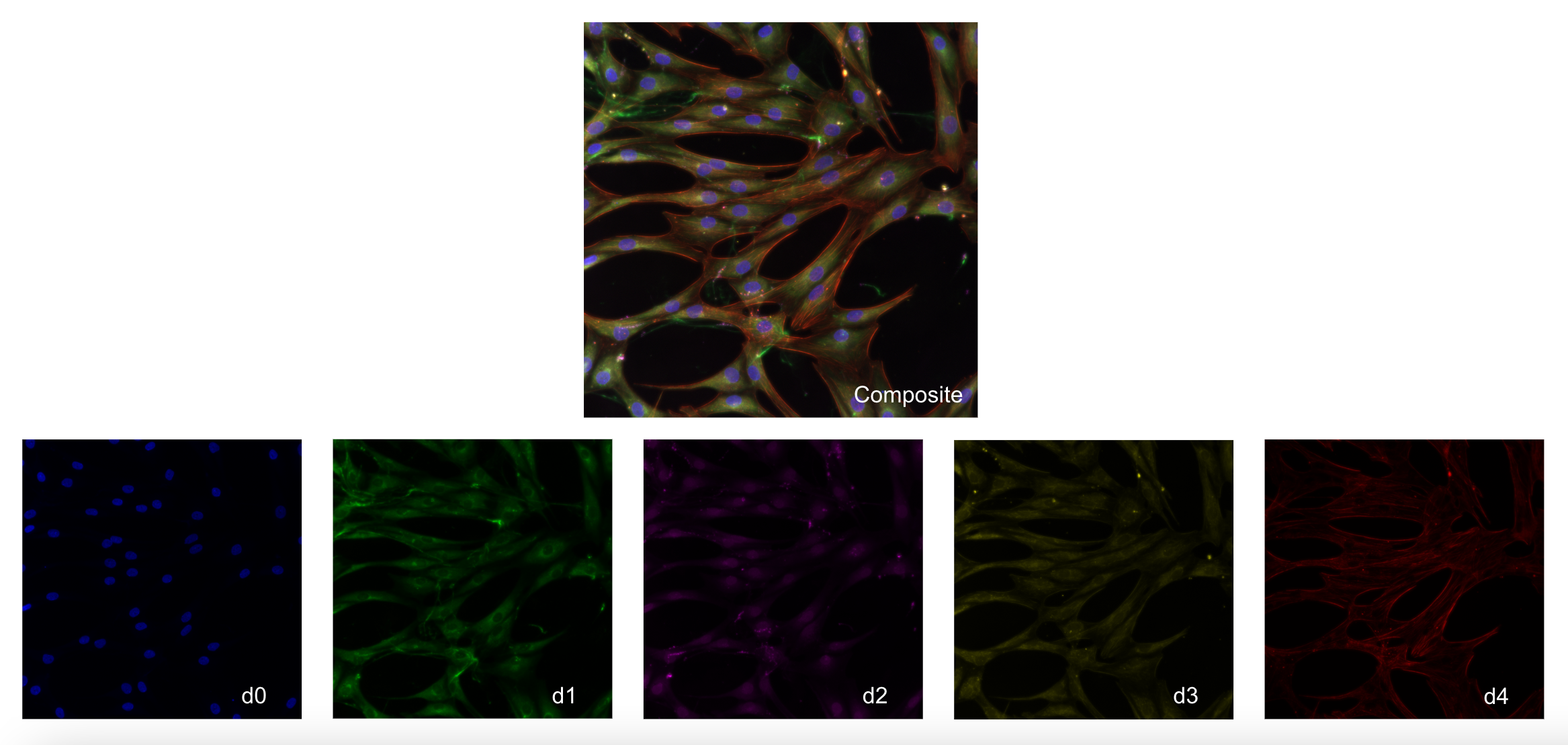
The data used in this project is a modified Cell Painting assay on cardiac fibroblasts from 3 patients that suffered from cardiac arrest.
In this modified Cell Painting, there are five channels:
d0(Nuclei)d1(Endoplasmic Reticulum)d2(Golgi/Plasma Membrane)d3(Mitochondria)d4(F-actin)
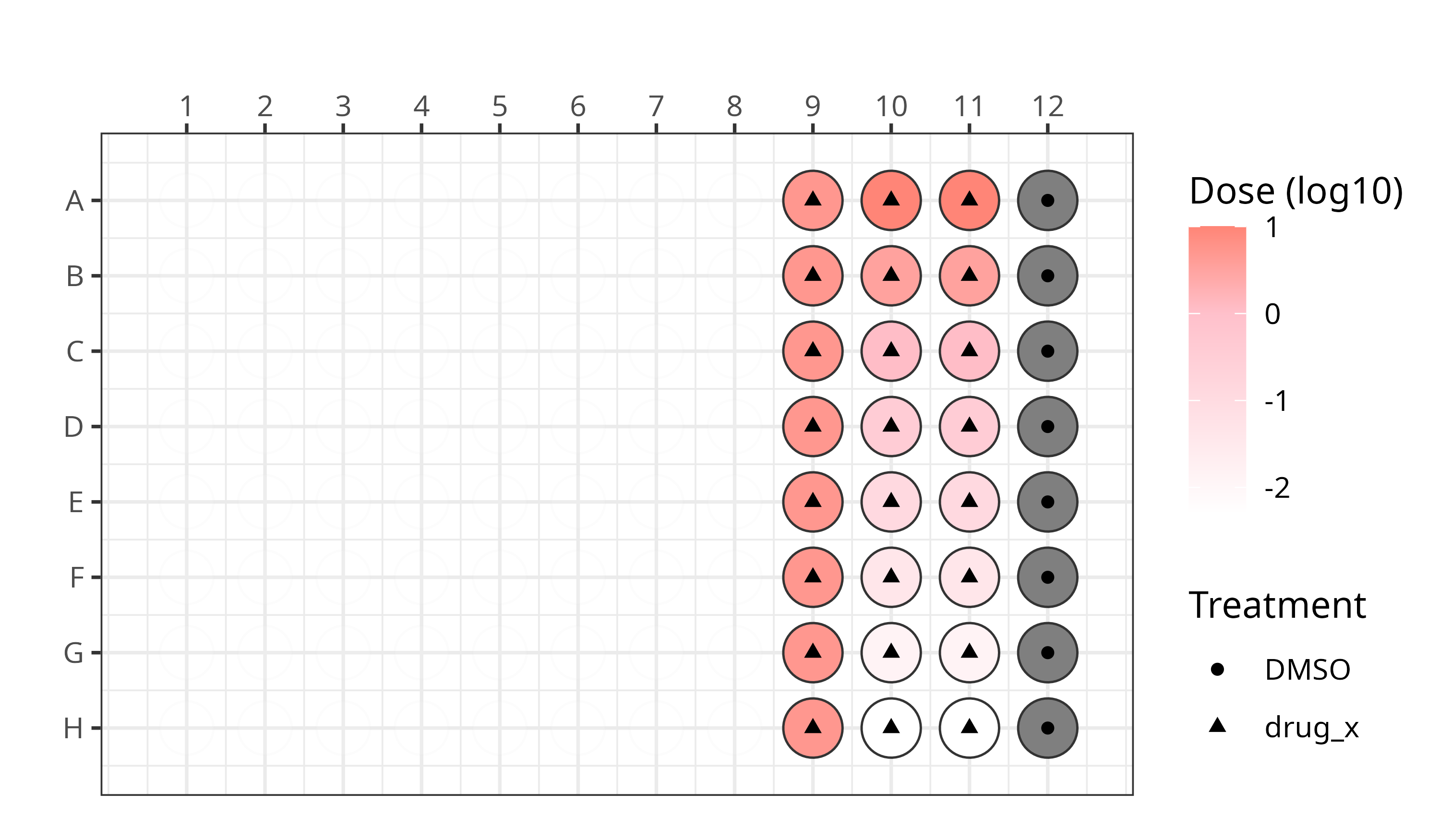
We applied this modified Cell Painting assay using the following plate design:
See our platemaps for more details.
The goals of this project are:
- To identify morphology features from cardiac fibroblasts that distinguish cardiac patients
- To discover a cell morphology biomarker associated with drug treatment to reverse fibrosis scarring caused by cardiac arrest.
| Module | Purpose | Description |
|---|---|---|
| 0.download-data | Download CFReT pilot data | Download pilot images for the CFReT project |
| 1.preprocessing-data | Perform Illumination Correction (IC) | Use BaSiCPy to perform IC on images per channel |
| 2.cellprofiler_processing | Apply feature extraction pipeline | Extract hundreds of morphology features per imaging channel |
| 3.process-cfret-features | Get morphology features analysis ready | Apply pycytominer to perform single cell normalization and feature selection |
| 4.analyze-data | Analyze the single cell profiles to achieve goals listed above | Several independent analyses to describe data and test hypotheses |