SpliCeAT: Integrated pipeline for detection and quantification of aberrant transcripts with novel splicing events
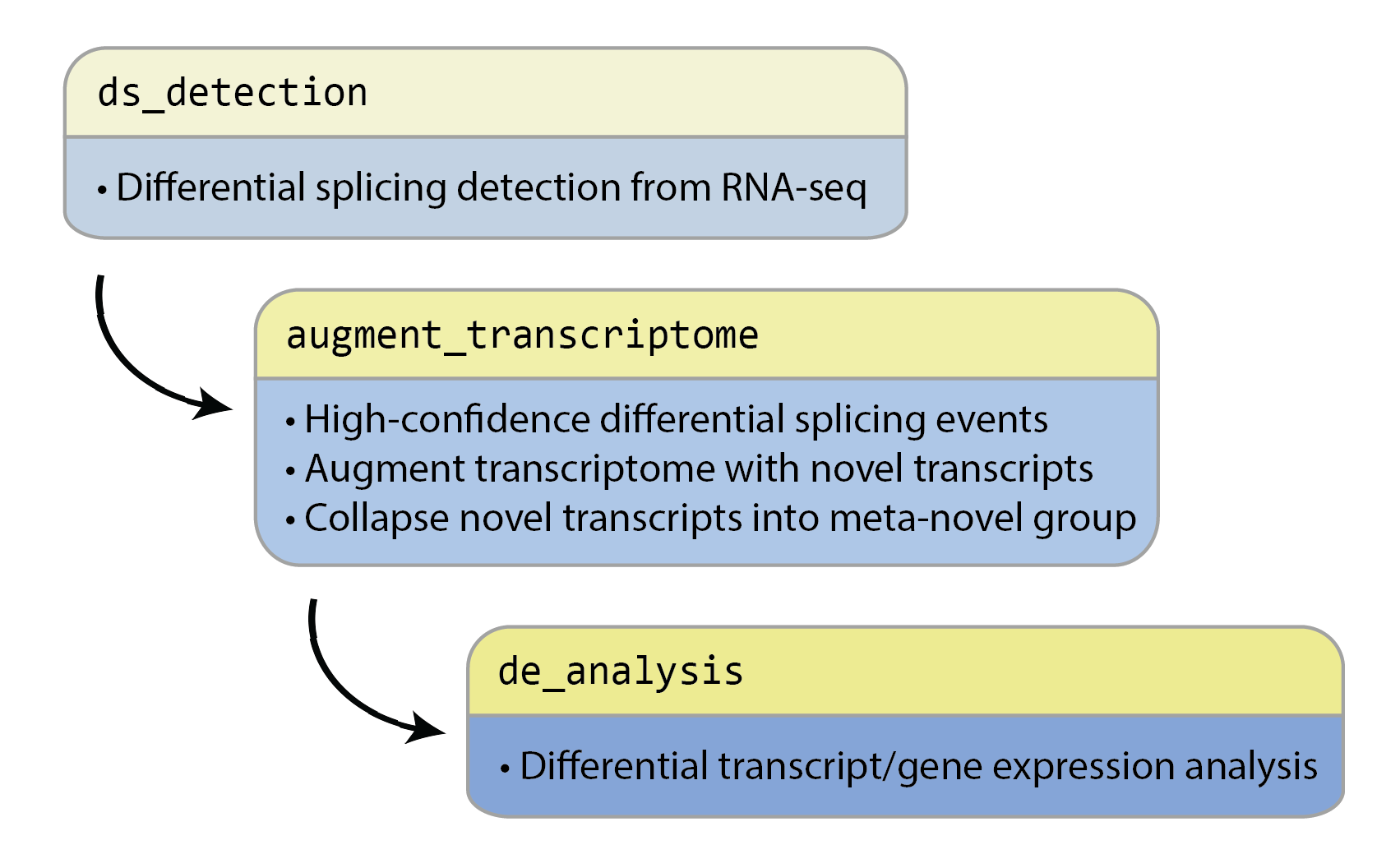
This repository contains the following Snakemake pipelines and scripts, to be run in this order:
- Differential splicing detection (
ds_detection) - Generation of augmented transcriptome (
augment_transcriptome) - Differential expression analysis (
de_analysis)
- FASTQ sample files of 2 conditions (control & treatment)
- Aligned BAM files using STAR, indexed using samtools 1
- Gene annotation (GTF & GFF3) in GENCODE format
Download the respective gene annotations and genome files for the species of interest and place them into a location of your choice:
- A
GFF3gene annotation file (e.g.gencode.vM29.primary_assembly.annotation.gff3). Unzip the file usinggunziptool. - A
GTFgene annotation file (e.g.gencode.vM29.primary_assembly.annotation.gtf.gz). - A
FASTAgenome file (e.g.GRCm39.primary_assembly.genome.fa.gz).
You may obtain the annotation files from Gencode (mouse).
git clone https://github.com/ys-lim/SpliCeAT.git
Footnotes
-
The pipelines expect RNA-seq alignments/BAM files to be labelled as
sample_Aligned.sortedByCoord.out.bam(as in STAR output format). Nevertheless, modifications can be made (at the user's discretion) in the Snakemake rules to account for alignments generated by other tools (e.g. HISAT2). ↩