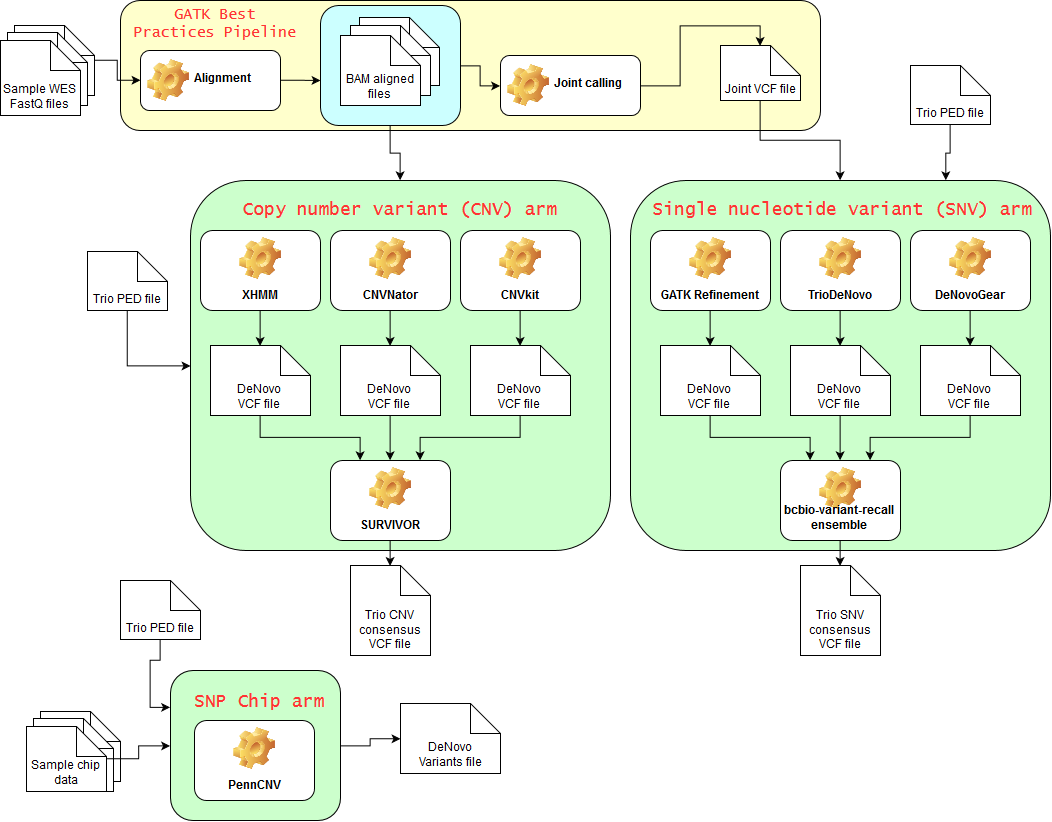
The goal in this project is to construct a pipeline for finding denovo mutations related to neurodevelopmental disorders using family study designs, similar to pipelines previously developed for cancer research. It combines different types of variants by multiple callers, and uses both chip and exome sequencing data.
The end goal is to compare annotated variants to public databases (dbVar, ClinVar), possibly indicating further sequencing or mechanistic avenues.
This is an ongoing project, and I've been journaling about my experiences in this project's GitHub wiki.
The current implementation needs the following files:
- Whole Exome Sequencing (WES) data for each participant
- Trio pedigree files (.ped), one trio per file
- SNP chip data for each participant
Note that some tools (e.g. XHMM, GATK joint calling) make use of all samples at the same time, regardless of their family structure. Also note that SNP chip data is not required, but you'll only be able to run 2 of the 3 arms in the pipeline. This approach of consensus calling and multiple sources of information tries to reduce false positives and increase a clinician's confidence on the results provided.
The end result of each analysis arm is a file containing de-novo mutations for each trio. The options for analysis using each result are many. For example:
- If some trios contain affected individuals and other don't, one option for filtering variants is to include only the ones that appear in most (or all) affected trios. Then, one can check how often those de-novo mutations appeared in the unnafected trios.
- If affected and unnafected trios come from the same family, this variant comparison can be performed within family first, before checking across families.
- The parameters specific to each caller can be tweaked to increase/decrease sensitivity and specificity, and hence obtain more/less variants in the final consensus call of each arm.
- Compare annotated variants to public databases, possibly indicating further sequencing or mechanistic avenues.
The one thing all arms will need is one .ped file for each trio. I like the convention FAMID_trioX.ped, and always making the trio with the affected child trio1. So, for example, 9005_trio1.ped and 9005_trio2.ped represent 4 people. The parents are the same in both pedigrees, and the child in 9005_trio1.ped is affected. For details on the .ped file, please see this. All pedigree files should be headless.
The SNP Chip pipeline can be run in paralle with the other two, but SNV and CNV need the files from GATK Best Practices pipeline. So, we'll start by running that.
First, modify the script gatk_upToSingleCalls.sh to reflect your environment. This script runs the GATK pipeline up to calls for the single sample. It takes the sample name, assuming all variables are properly set. I highly recommend running it in a computer cluster. In my case, we have a cluster running SLURM, so I swarm the jobs this way:
while read s; do echo "bash ~/autodenovo/gatk_upToSingleCalls.sh $s" >> swarm.single; done < sample_ids.txt
swarm -f swarm.single -t 16 -g 55 --job-name gatk_single --logdir trash --time=48:00:00 --gres=lscratch:100where sample_ids.txt is a file with one sample name per line. This should take about 1-2 days to run per participant, so there's the beauty of running them all in parallel.
We can check which samples didn't finish properly:
while read s; do if [ ! -e VCF/${s}/${s}.g.vcf.idx ]; then echo $s; fi; done < sample_ids.txtAnd then go through the logs to see why/where they failed.
All tools in the SNV arm need VCF files, preferably joint called. So, we run the second stage of the GATK Best practices pipeline, in which we perform joint calling:
bash ~/autodenovo/gatk_jointCalling.sh /data/NCR_SBRB/big_fake_simplex/sample_ids.txtAll these tools can be run in parallel, as they only depend on the jointly called VCF. If we don't do any sort of pre-filtering, it's simple:
mkdir triodenovo
cd triodenovo
while read t; do ../../software/triodenovo.0.05/bin/triodenovo --ped ../${t}.ped --in_vcf ../VCF/recalibrated_variants.vcf --out ${t}_denovo.vcf; done < ../trio_ids.txtAnd fix an error in the VCF header:
while read t; do
sed -e "s/Type\=Denovo\ Quality/Type=Float/g" ${t}_denovo.vcf > ${t}_denovo_v2.vcf
done < ../trio_ids.txtmkdir dng
cd dng
while read t; do
../../software/denovogear-v1.1.1-Linux-x86_64/bin/dng dnm auto --ped ../${t}.ped --output_vcf ${t}_dnm.vcf --vcf ../VCF/recalibrated_variants.vcf; done < ../trio_ids.txtwhile read s; do echo "bash ~/autodenovo/gatk_refine.sh $s" >> swarm.refine; done < trio_ids.txt
swarm -f swarm.refine -t 2 -g 55 --job-name gatkr --logdir trash --time=48:00:00 --gres=lscratch:100And, if we also want to create unfiltered calls to better compare with the other tools, after the processed from above finish running we do:
while read s; do echo "bash ~/autodenovo/gatk_refine_noFilter.sh $s" >> swarm.refineNF; done < trio_ids.txt
swarm -f swarm.refineNF -t 2 -g 55 --job-name gatkr --logdir trash --time=48:00:00 --gres=lscratch:100Now we have one file with results for each trio, for each tool. Time to ensemble the calls:
module load bcbio-nextgen
ref_fa=/fdb/igenomes/Homo_sapiens/UCSC/hg19/Sequence/WholeGenomeFasta/genome.fa;
while read t; do
echo Ensembling $t
bcbio-variation-recall ensemble --numpass 3 --names GATK,DeNovoGear,TrioDeNovo ${t}_ensemble.vcf $ref_fa gatk_refine/${t}_hiConfDeNovo.vcf dng/${t}_dnm.vcf triodenovo/${t}_denovo_v2.vcf;
gunzip ${t}_ensemble.vcf.gz;
rm -rf ${t}_ensemble-work ${t}_ensemble.vcf.gz.tbi;
done < trio_ids.txt
mkdir snv_arm
mv *_ensemble.vcf snv_armIn my case, I have nuclear families (parents and siblings) where only one kid is affects. So, I'll look for denovo variants that appear in the affected sibling, but not in the unaffected one(s). Spit those SNVs to a file:
cd snv_arm
for fam in `ls *_trio1_ensemble.vcf | sed -e 's/_trio1_ensemble\.vcf//g' -`; do
ntrios=`ls -1 ${fam}_trio?_ensemble.vcf | wc -l`;
if [ $ntrios -gt 1 ]; then
echo $fam;
cut -f 1,2 ${fam}_trio1_ensemble.vcf | grep -v '#' - > ${fam}_possible_snvs.txt;
cat ${fam}_trio[2..$ntrios]_ensemble.vcf > ${fam}_control_snvs.txt;
while read snv; do
if ! grep -q "$snv" ${fam}_control_snvs.txt; then
echo $snv >> interesting_snvs.txt;
fi;
done < ${fam}_possible_snvs.txt;
fi;
doneFinally, count how often each structural variant appears in the file:
sort interesting_snvs.txt | uniq -cdUnfortunately, in my case the most times the SNVs happened were 2, which is not necessarily exciting as I have 21 affected families. One option is to make filters less conservative, or require only 2 or 1 tools to call the variant. So, good luck!
This takes a while, so we swarm it:
while read s; do echo "bash ~/autodenovo/gatk_refine.sh $s" >> swarm.refine; done < trio_ids.txt
swarm -f swarm.refine -t 2 -g 55 --job-name gatkr --logdir trash --time=48:00:00 --gres=lscratch:100After we ran the first part of the GATK Best Practices pipeline (variant calling for single samples), we should have aligned BAMs for all our samples. For this arm, we don't need to wait for the joint calling steps, as we'll be working directly from BAMs. Also, all tools can be run in parallel:
mkdir xhmm
while read s; do echo "bash ~/autodenovo/xhmm_get_DOC.sh $s" >> xhmm/swarm.DOC; done < sample_ids.txt
cd xhmm
swarm -f swarm.DOC -t 4 -g 55 --job-name xhmmDOC --logdir trash --time=48:00:00 --gres=lscratch:100This one doens't take very long, so we don't need to swarm it:
cp /usr/local/apps/XHMM/2016-01-04/params.txt .
bash ~/autodenovo/xhmm_eval.shAutodenovo includes a script to run all steps in CNVnator, so it's quite easy to just try it for a few different window options:
mkdir cnvnator
cd cnvnator
while read s; do
for w in 30 100 500; do
echo "bash ~/autodenovo/cnvnator_full.sh $s $w" >> swarm.cnvnator;
done;
done < ../sample_ids.txt
swarm -f swarm.cnvnator -t 2 -g 16 --job-name cnvnator --logdir trash -m cnvnator --gres=lscratch:50 --time=48:00:00Running CNVkit is somewhat automated as well. First thing, create the access files for each of the windows you'll be playing with:
mkdir cnvkit
cd cnvkit
for w in 1 5 10 50 100; do
/data/NCR_SBRB/software/cnvkit/cnvkit.py access /fdb/igenomes/Homo_sapiens/UCSC/hg19/Sequence/WholeGenomeFasta/genome.fa -s ${w}000 -o access-${w}kb.hg19.bed;
doneNow, swarm all samples, for the different windows:
while read s; do for w in 1 5 10 50 100; do echo "bash ~/autodenovo/cnvkit_full.sh $s $w" >> swarm.cnvkit; done; done < ../sample_ids.txt
swarm -f swarm.cnvkit -t 6 -g 16 --job-name cnvkit --logdir trash --time=48:00:00 --gres=lscratch:10As the SNP chip arm doesn't depend on the exome data, let's run it while we wait for the GATK pipeline results.
I strongly advise you to look at the PennCNV documentation to figure out how to prepare your input files.
In my case, we used Illumina's InfiniumExome-24_v1.0 chips. So, I used GenomeStudio (free to download, but it does require Windows) to export a Final Report with the columns we need (SNP Name, Sample ID, Allele1 - Top, Allele2 - Top, GC Score, Log R Ratio, B Allele Freq.).
We also have a few samples that were genotyped on Illumina's HumanExome-12v1-2. Louckily they are all in the same family, otherwise you'd have to come up with files representing the intersection of the array. Contact me if you'd like instructions on how to do that, or check the Wiki, because I had to do it for the sample data I was using there.
For now, let's go ahead and split the data:
mkdir penncnv
cd penncnv
module load penncnv
mkdir HumanExome
split_illumina_report.pl -prefix HumanExome/ fs_ccgo_box4_FinalReport.txt
mkdir InfiniumExome
split_illumina_report.pl -prefix InfiniumExome/ newBox225_FinalReport.txt
split_illumina_report.pl -prefix InfiniumExome/ newBox226_FinalReport.txt We then have to download PFB files from Illumina's website. They don't have those exact files there, but all we're looking for is the Population Frequency of B Allele, which is calculated in their MAF files. We just need to extract the column that corresponds to our population. In my case, I'll get this and this. So, to make our PFB files, we need:
# get the first 4 columns, and ignore the first line
cut -f 1,2,3,4 HumanExome-12v1-2_A_MAF.txt | tail -n +2 > HumanExome.pfb
cut -f 1,2,3,4 Population\ Reports\ \(MAF\,\ Copy\ Numbers\)/InfiniumExome-24v1-0_A1_PopulationReport_MAF.txt | tail -n +2 > InfiniumExome.pfbI'm going to use the default HMM parameters, but feel free to play with them if you want to increase/decrease sensitivity and specificity. Finally, I also created a GC content file to test the corrections implemented in PennCNV. There's a simple script to do it, but you'll likely need to change it to fit your needs. Also, this takes a few hours to run, so use it wisely.
bash ~/autodenovo/penncnv_create_gcmodel.sh HumanExome.pfb /fdb/igenomes/Homo_sapiens/UCSC/hg19/Sequence/WholeGenomeFasta/genome.fa HumanExome.h19.gcmodel
bash ~/autodenovo/penncnv_create_gcmodel.sh InfiniumExome.pfb /fdb/igenomes/Homo_sapiens/UCSC/hg19/Sequence/WholeGenomeFasta/genome.fa InfiniumExome.h19.gcmodelNow that all files are ready, there's a handy script to run PennCNV to find denovo mutations. Because it runs quite quickly, we can loop around it for all our trio pedigrees:
for t in `ls -1 *trio*ped | sed -e 's/\.ped//'`; do
bash ~/autodenovo/penncnv_run_trio.sh $t InfiniumExome.pfb InfiniumExome.hg19.gcmodel;
done
# redo it for the family in a different box
for t in `ls -1 10369_trio*ped | sed -e 's/\.ped//'`; do
bash ~/autodenovo/penncnv_run_trio.sh $t HumanExome.pfb HumanExome.hg19.gcmodel;
endAssuming that all our trios have been coded as FAMID_trioX.ped, as above.
The last step is to figure out which CNVs are de novo in each trio. That's simply the CNVs in the offspring that are not in the parents:
for trio in `ls -1 *trio*ped | sed -e 's/\.ped//'`; do
echo $trio >> penncnv_results.txt
grep mother penncnv/results/${trio}.triocnv > mom_snps;
grep father penncnv/results/${trio}.triocnv > dad_snps;
cat mom_snps dad_snps > parent_snps;
for snp in `grep offspring penncnv/results/${trio}.triocnv | cut -d' ' -f 1`; do
if ! grep -q $snp parent_snps; then
echo "De Novo CNV: $snp" >> penncnv_results.txt
fi;
done
rm *_snps;
done
for trio in `ls -1 *trio*ped | sed -e 's/\.ped//'`; do
echo $trio >> penncnv_adjusted_results.txt
grep mother penncnv/results/${trio}.adjusted.triocnv > mom_snps;
grep father penncnv/results/${trio}.adjusted.triocnv > dad_snps;
cat mom_snps dad_snps > parent_snps;
for snp in `grep offspring penncnv/results/${trio}.adjusted.triocnv | cut -d' ' -f 1`; do
if ! grep -q $snp parent_snps; then
echo "De Novo CNV: $snp" >> penncnv_adjusted_results.txt
fi;
done
rm *_snps;
done