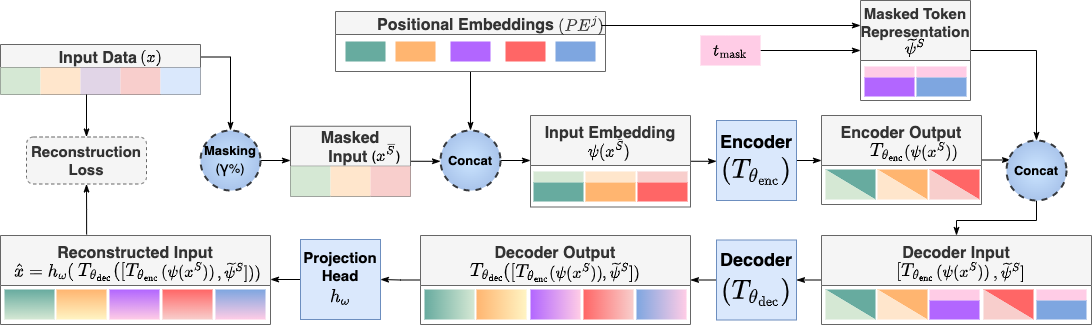
This repository is the official implementation of MET.
Disclaimer : This is not an officially supported Google product.
To run experiments mentioned in the paper and install requirements use python version >=3.7:
git clone http://github.com/google-research/met
cd met
pip install -r requirements.txt
To train the MET-S model mentioned in the paper (model without adversarial training step) for FashionMNIST dataset, run this command:
python3 train.py
The following hyper-parameters are available for train.py :
- embed_dim : Embedding dimension
- ff_dim : Feed-Forward dimension
- num_heads : Number of heads
- model_depth_enc : Depth of Encoder/ Number of transformers in Encoder stack
- model_depth_dec : Depth of Decoder/ Number of transformers in Decoder stack
- mask_pct : Masking Percentage
- lr : Learning rate
Each of the above can be changed by adding --flag_name=flag_value to train.py. For example :
python3 train.py --model_depth_enc=1
The model is saved here by default
To train the MET model in the paper for FashionMNIST dataset trained using Adversarial training, run this command:
python3 train_adv.py
The following hyper-parameters are available for train.py :
- embed_dim : Embedding dimension
- ff_dim : Feed-Forward dimension
- num_heads : Number of heads
- model_depth_enc : Depth of Encoder/ Number of transformers in Encoder stack
- model_depth_dec : Depth of Decoder/ Number of transformers in Decoder stack
- mask_pct : Masking Percentage
- lr : Learning rate
- radius : Radius of L2 norm ball around the input data point
- adv_steps : Adversarial loop length
- lr_adv : Adversarial Learning Rate
Each of the above can be changed by adding --flag_name=flag_value to train.py. For example :
python3 train_adv.py --radius=14
The model is saved here by default
You can try using the model on any new dataset by creating a csv file. The first column of the csv file should be class followed by the attributes. Sample csv files are available in data
To pass on the csv file to any of the training and evaluation scripts use the following flags :
- num_classes : Number of classes
- model_kw : Keyword for model (Eg fmnist for fashion-mnist)
- train_len : Length of train csv
- train_data_path : Path to train csv file
- test_len : Length of test csv
- test_data_path : Path to test csv files
- By default models are stored in saved_models. You can change the training path using flag model_path.
- Synthetic dataset can be created using get_2d_dataset.py. By default a created dataset is available in data
Pretrained models for FashionMNIST for optimal adversarial training setting is available in saved_models. You can extract the models using command:
7z e fmnist_saved.7z.001
7z e fmnist_saved_adv.7z.001
To evaluate the saved MET-S model run
python3 eval.py --model_path="./saved_models/fmnist_64_1_64_6_1_70_1e-05" --model_path_linear="./saved_models/fmnist_linear_64_1_64_6_1_70_1e-05"
To evaluate the saved MET model run
python3 eval.py --model_path="./saved_models/fmnist_adv_64_1_64_6_1_70_1e-05" --model_path_linear="./saved_models/fmnist_linear_adv_64_1_64_6_1_70_1e-05"
By default results are written to met.csv.
| Type | Methods | FMNIST | CIFAR10 | MNIST | CovType | Income |
|---|---|---|---|---|---|---|
| Supervised Baseline | MLP | 87.57 ± 0.13 | 16.47 ± 0.23 | 96.98 ± 0.1 | 65.45 ± 0.09 | 84.35 ± 0.11 |
| RF | 87.19 ± 0.09 | 36.75 ± 0.17 | 97.62 ± 0.18 | 64.94 ± 0.12 | 84.6 ± 0.2 | |
| GBDT | 88.71 ± 0.07 | 45.7 ± 0.27 | 100 ± 0.0 | 72.96 ± 0.11 | 86.01 ± 0.06 | |
| RF-G | 89.84 ± 0.08 | 29.28 ± 0.16 | 97.63 ± 0.03 | 71.53 ± 0.06 | 85.57 ± 0.13 | |
| MET-R | 88.81 ± 0.12 | 28.97 ± 0.08 | 97.43 ± 0.02 | 69.68 ± 0.07 | 75.50 ± 0.04 | |
| Self-Supervised Methods | VIME | 80.36 ± 0.02 | 34 ± 0.5 | 95.74 ± 0.03 | 62.78 ± 0.02 | 85.99 ± 0.04 |
| DACL+ | 81.38 ± 0.03 | 39.7 ± 0.06 | 91.35 ± 0.075 | 64.17 ± 0.12 | 84.46 ± 0.03 | |
| SubTab | 87.58 ± 0.03 | 39.32 ± 0.04 | 98.31 ± 0.06 | 42.36 ± 0.03 | 84.41 ± 0.06 | |
| Our Method | MET-S | 90.90 ± 0.06 | 47.96 ± 0.1 | 98.98 ± 0.05 | 74.13 ± 0.04 | 86.17 ± 0.08 |
| MET | 91.68 ± 0.12 | 47.92 ± 0.13 | 99.17+-0.04 | 76.68 ± 0.12 | 86.21 ± 0.05 |
| Datasets | Metric | MLP | RF | GBDT | RF-G | MET-R | DACL+ | VIME | SubTab | MET |
|---|---|---|---|---|---|---|---|---|---|---|
| Obesity | Accuracy | 58.1 ± 0.07 | 65.99 ± 0.12 | 67.19 ± 0.04 | 58.39 ± 0.17 | 58.8 ± 0.59 | 62.34 ± 0.12 | 59.23 ± 0.17 | 67.48 ± 0.03 | 74.38 ± 0.13 |
| AUROC | 52.3 ± 0.12 | 64.36 ± 0.07 | 64.4 ± 0.05 | 54.45 ± 0.08 | 53.2 ± 0.18 | 61.18 ± 0.07 | 57.27 ± 0.21 | 64.92 ± 0.06 | 71.84 ± 0.15 | |
| Income | Accuracy | 84.35 ± 0.11 | 84.6 ± 0.2 | 86.01 ± 0.06 | 85.57 ± 0.13 | 75.50 ± 0.04 | 85.99 ± 0.24 | 84.46 ± 0.03 | 84.41 ± 0.06 | 86.21 ± 0.05 |
| AUROC | 89.39 ± 0.2 | 91.53 ± 0.32 | 92.5 ± 0.08 | 90.09 ± 0.57 | 83.48 ± 0.23 | 89.01 ± 0.4 | 87.37 ± 0.07 | 88.95 ± 0.19 | 93.85 ± 0.33 | |
| Criteo | Accuracy | 74.28 ± 0.32 | 71.09 ± 0.05 | 72.03 ± 0.03 | 74.62 ± 0.08 | 73.57 ± 0.12 | 69.82 ± 0.06 | 68.78 ± 0.13 | 73.02 ± 0.08 | 78.49 ± 0.05 |
| AUROC | 79.82 ± 0.17 | 77.57 ± 0.1 | 78.77 ± 0.04 | 80.32 ± 0.16 | 79.17 ± 0.17 | 75.32 ± 0.27 | 74.28 ± 0.39 | 76.57 ± 0.05 | 86.17 ± 0.2 | |
| Arrhythmia | Accuracy | 59.7 ± 0.02 | 68.18 ± 0.02 | 69.79 ± 0.12 | 60.6 ± 0.05 | 51.67 ± 0.1 | 57.81 ± 0.47 | 56.06 ± 0.04 | 60.1 ± 0.1 | 81.25 ± 0.12 |
| AUROC | 72.23 ± 0.06 | 90.63 ± 0.08 | 92.19 ± 0.05 | 74.02 ± 0.12 | 58.36 ± 0.32 | 69.23 ± 0.98 | 67.03 ± 0.27 | 69.97 ± 0.07 | 98.75 ± 0.04 | |
| Thyroid | Accuracy | 50 ± 0.0 | 94.94 ± 0.1 | 96.44 ± 0.07 | 50 ± 0.0 | 57.42 ± 0.37 | 60.03 ± 0.05 | 66.1 ± 0.19 | 59.9 ± 0.16 | 98.1 ± 0.08 |
| AUROC | 62.3 ± 0.12 | 99.62 ± 0.03 | 99.34 ± 0.02 | 52.65 ± 0.13 | 82.03 ± 0.26 | 86.63 ± 0.1 | 94.87 ± 0.03 | 88.93 ± 0.12 | 99.81 ± 0.09 |