Repository for Palmer amaranth detection in cotton at different growth stages. It accompanies the publication in Computers and Electronics in Agriculture: Multi-growth stage plant recognition: a case study of Palmer amaranth (Amaranthus palmeri) in cotton (Gossypium hirsutum) available here. Please consider citing this preprint if you use the work in your research.
Model training is completed using Google Colab, and as such image and annotation data needs to be stored on your Google Drive. Follow the steps below to set up the data and repository for replication of the training procedure used.
Clone the repository into your Google Drive by mounting Drive, navigating to it and running:
git clone https://github.com/geezacoleman/Palmer-detection palmer-amaranth
Download the complete PAGS8 dataset from Weed-AI and unzip the images into the data/images directory.
Organise the data into respective splits and folds using the splits in data/splits and the labels for each class
grouping under data/labels_GROUP. Make sure you are in the palmer-amaranth directory within your Google Drive first.
Run the utility function:
python setup_dataset.py --data-dir 'data' --output-dir 'datasets'
Change the flags to your custom locations as needed. The defaults will function correctly from the root directory of this repository.
A tool to check annotations for YOLO and PASCAL VOC (XML) format has been developed. Using utils.py, simply provide
the path to the image and labels directory and the class names.
if __name__ == "__main__":
classes = ['PA-1', 'PA-2', 'PA-3', 'PA-4', 'PA-5', 'SPA-1', 'SPA-2', 'SPA-3']
visualise(label_dir='data/xml',
images_dir=r'data/images',
classes=classes)Update these variables as necessary.

Screenshot of the annotation checker. Use 'space' to move to the next image and 'esc' to exit.
python setup_dataset.py
This may take some time as it copies the dataset five times into respective folds and splits.
Once you have finished setting the repository up, you are ready to start training models. Model training was conducted using Google Colab Pro+. All training notebooks are available below.
| YOLO version | Google Colab Link |
|---|---|
| v3 | |
| v5 | |
| v6 | |
| v6 3.0 | |
| v7 | |
| v8 | |
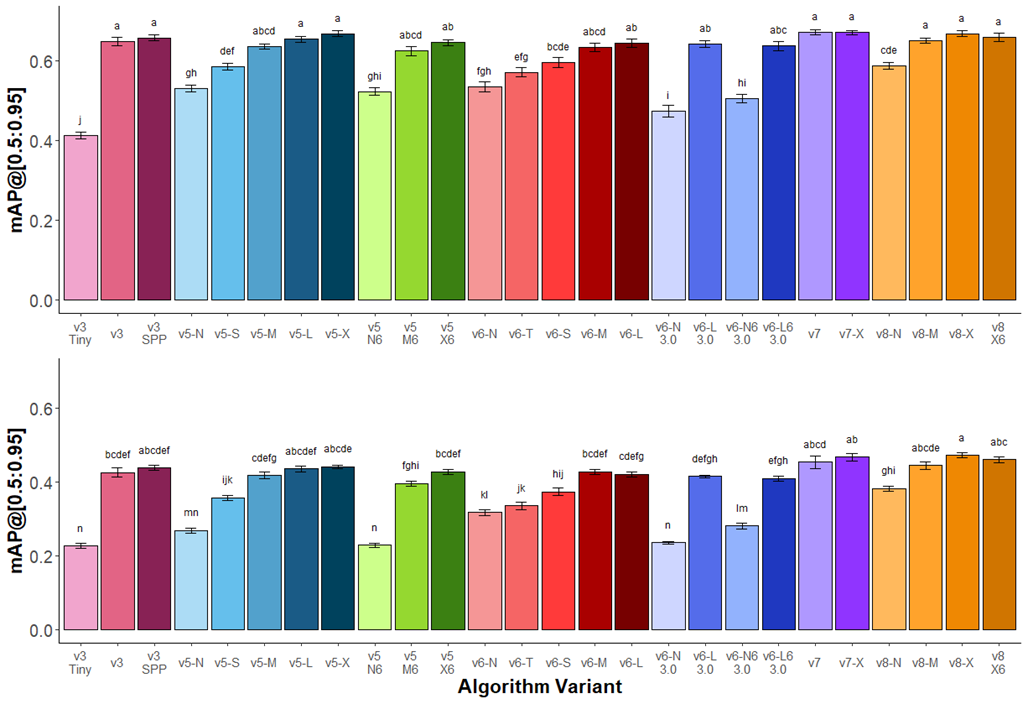
Figure 2 growth stage recognition performance of Amaranthus palmeri across YOLO versions and variants
trained on a single (top) class and all eight (bottom) classes. Variants within the same YOLO version are indicated by
similar colors. The largest variants within each series performed similarly. Standard error (n=5) of the mean is
provided as error bars. Tukey’s HSD lettering indicates significance (P < 0.05) between different variants within the single or eight class
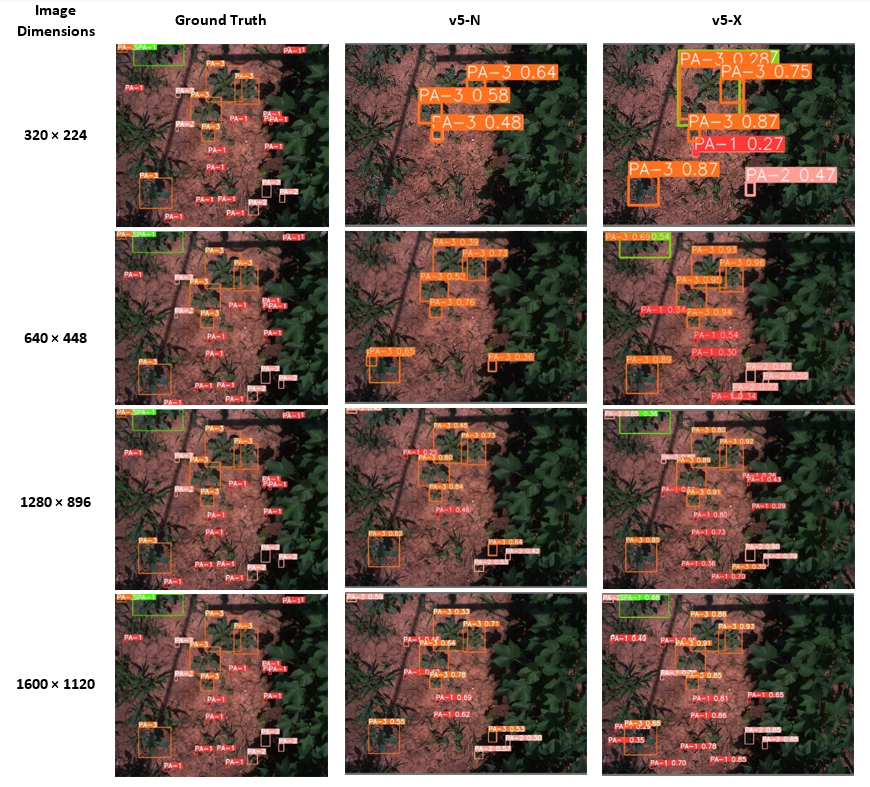
Figure 7 Illustration of growth stage detection at the four resolutions tested for the ground truth (left),
v5-X, and v5-N model variants. At lower resolutions, the difference between the smaller and larger model becomes apparent.
Please cite our work as:
Coleman GRY, Kutugata M, Walsh MJ, Bagavathiannan MV. Multi-growth stage plant recognition: A case study of Palmer amaranth (Amaranthus palmeri) in cotton (Gossypium hirsutum). Computers and Electronics in Agriculture 2024;217:108622. https://doi.org/10.1016/j.compag.2024.108622.
@article{coleman2024multi,
title={Multi-growth stage plant recognition: A case study of Palmer amaranth (\textit{Amaranthus palmeri}) in cotton (\textit{Gossypium hirsutum})},
author={Coleman, Guy R.Y. and Kutugata, Matthew and Walsh, Michael J. and Bagavathiannan, Muthukumar V.},
journal={Computers and Electronics in Agriculture},
volume={217},
pages={108622},
year={2024},
issn={0168-1699},
doi={10.1016/j.compag.2024.108622},
url={https://www.sciencedirect.com/science/article/pii/S0168169924000139},
keywords={High throughput phenotyping; Deep learning; Precision weed management; Computer vision; Open-access dataset, Benchmark}
}
This research made use of the open-source, YOLO family of object detection models. Repository clones made at the time the research was conducted are provided here for replication purposes. Only minor modifications have been made to the code. Please credit the original authors of these repositories listed below: