Dept. of Clinical and Experimental Biomedical Sciences "Mario Serio", University of Florence, Florence
Disentangling the human gut microbiota composition is a pre-requisite to unveil its involvement
in physiological and pathological host states. The human gut microbiota is composed by
Prokaryotes (Archaea and Eubacteria), Viruses and small Eukaryotes such as Fungi and Protista.
Over the last two decades, analysis of 16S DNA by metabarcoding allowed the convenient and
effective investigation of the gut Bacteria populations, unveiling several insights about their
compositional and functional features [1]. By contrast, only in recent years the interest in the
Eukaryotic microorganisms of the human gut microbiota has begun to emerge.
Aim of this work is therefore to deepen the knowledge about the Eukaryotic community of the
gut microbiota, with a particular focus on mycobiota, exploiting the metabarcoding data of
healthy control samples in publicly available NCBI BioProjects. For this investigation the
(Internal Transcribed Spacer) ITS1 DNA barcode was selected because of its superior reliability
in comparison with ITS2 [2]. Then, a compositional profile was obtained. Also, a focus about
KEGG [3] pathways potentially associated with investigated microbiota was performed.
Five NCBI BioProjects were selected by combining an in-house developed Python script and a
manual research on the NBCI BioProject website. The inclusion criteria were the presence of
healthy control in the study and the use of ITS1 sequence as investigated barcode. Then,
metabarcoding analysis was conducted on ITSoneWB * [4] a Galaxy workbench environment
based on the employment of ITSoneDB [5] as reference database for ITS1 sequences. The
workflow selected for the analysis was BioMaS [6]. In brief, BioMaS is an OTU (Operational
Taxonomic Unit) and ASV (Amplicon Sequence Variants) free method. After quality check and
trimming of reads it attempts to merge the paired end reads. Then, the ensemble of successfully
merged consensus, not merged and unpaired reads (obtained from successful, failed merging
and trimming, respectively) were aligned against reference sequences. The not merged and
unpaired reads are required to avoid loss of biodiversity information. Because of the high ITS1
length variability (i.e. 100-1000nt) merging fails can occur, generating not-merged and unpaired
sequences that are discarded by OTU- and ASV-based methods. Alignment results were used to
perform taxonomic classification.
Thus, data were cleaned by dietary or environment contaminants. Samples were rarefied to
uniform sequencing depths. Relative abundances were collapsed at genus and phylum levels.
Furthermore, a Chi-square test (FDR < 0.05) to assess the statistically significant taxa prevalent
among the samples was carried out.
KEGG pathways ontology analysis related to genera found in the samples was performed using
the following strategy. KEGG provides pathway tables for several fungal and protista species. By
using KEGG API, tables of species belonging to genera found in our samples were locally
downloaded, when available. Then, for each genus the common pathways among species were
selected as genus associated. Genera associated pathways significantly prevalent among
samples were selected (Chi-square, FDR < 0.01).
Finally, a gender specific analysis for α-diversity was performed within a subset of samples for
which sex metadata were publicly available.
Five studies were selected according to the inclusion criteria described above. The NCBI
BioProject accession numbers are PRJNA778607, PRJNA607176, PRJNA317653, PRJEB25916,
PRJEB53019 with 42, 47, 10, 61, 11 healthy control subjects, respectively. The total amount of
healthy controls was 171 samples. After rarefaction, 60 samples were retained. The prevalent
found genera (FDR < 0.05) were Candida, Ascomycota, Penicillium, Cryptococcus,
Basidiomycota, Aspergillus, Fusarium, Geotrichum, Cladosporium, Pichia, Saccharomyces,
Galactomyces, Debaryomyces, Filobasidium, Alternaria. At phylum level Ascomycota,
Basidiomycota and Mucoromycota were found.
From the 131 statistically prevalent pathways 7 degradative, 26 biosynthetic and 50 metabolic
were found, including Atrazine degradation, biosynthesys of wax and Folate, Caffeine
metabolism, vesicular transport, ABC transporters and beta-Lactam resistance. Intriguingly,
biosynthesis of antibiotics such as Carbapenem, Monobactam, Penicillin, Cephalosporin were
found.
Furthermore, a subset of samples reporting sex metadata was selected to perform gender-
specific α-diversity consideration. According to the publicly available metadata of samples, the
female group was composed by 12, while the male group 31 samples. The Wilcoxon test
comparing male vs female distributions for Inverse Simpson resulted statistically significant (p-
value 0.011).
In conclusion, this study highlights the potential of publicly available metabarcoding data reuse
for the exploration of human gut eukaryotic microorganisms composition and their associatedbiochemical pathways.
[1] P. J. Turnbaugh, R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, e J. I. Gordon, «The
Human Microbiome Project», Nature, vol. 449, fasc. 7164, Art. fasc. 7164, ott. 2007, doi:
10.1038/nature06244.
[2] X.-C. Wang et al., «ITS1: a DNA barcode better than ITS2 in eukaryotes?», Mol. Ecol.
Resour., vol. 15, fasc. 3, Art. fasc. 3, 2015, doi: 10.1111/1755-0998.12325.
[3] M. Kanehisa e S. Goto, «KEGG: Kyoto Encyclopedia of Genes and Genomes», Nucleic Acids
Res., vol. 28, fasc. 1, pp. 27–30, gen. 2000.
[4] M. A. Tangaro et al., «ITSoneWB: profiling global taxonomic diversity of eukaryotic
communities on Galaxy», Bioinformatics, fasc. btab431, giu. 2021, doi:10.1093/bioinformatics/btab431.
[5] M. Santamaria et al., «ITSoneDB: a comprehensive collection of eukaryotic ribosomal RNA
Internal Transcribed Spacer 1 (ITS1) sequences», Nucleic Acids Res., vol. 46, fasc. D1, Art. fasc.
D1, gen. 2018, doi: 10.1093/nar/gkx855.
[6] B. Fosso et al., «BioMaS: a modular pipeline for Bioinformatic analysis of Metagenomic
AmpliconS», BMC Bioinformatics, vol. 16, fasc. 1, Art. fasc. 1, dic. 2015, doi: 10.1186/s12859-
015-0595-z.
We would like to tank ITSoneWB mantainers Bruno Fosso at University of Bari and Marco Tangaro at IBIOM National Research Council for their support in the ITSoneWB usage.
In this study, KEGG pathways representation is obtained relying on genera composition.
For each genus found in our samples, the script kegg_pathways_analysis.py
interrogates KEGG database API (https://rest.kegg.jp/list/pathway/),
downloading pathways of species belonging to genera found in samples. Then, for a reliable
genus representation of KEGG pathways, only pathways in common among
species of the same genus were reported.
The result is a DataFrame reporting KEGG pathways as rows and genera
as columns filled by 0/1 as boolean.
For more inspect the kegg_pathways_analysis.py code.
Prevalent genera among samples were found by using the following R script:
genera <- read.csv('./samples/BioMas_raref_genus_collapsed.csv', row.names =1)
g.list <- genera$genus[2:614]
g.list
rownames(genera) <- genera$genus
g.TF <- data.frame(row.names = g.list)
for (g in g.list){
g.TF[g, c('FALSE', 'TRUE')] <- table(genera[g,2:61] > 0)
tst <- prop.test(x=g.TF[g,'TRUE'],n= 60, alternative = 'greater')
g.TF[g, 'p.value'] <- tst$p.value
g.TF[g, 'prop'] <- tst$estimate[['p']]
}
g.TF$padj <- p.adjust(p = g.TF$p.value,
method = 'BH',
n = 613)
sign.gen <- g.TF[g.TF$padj < 0.05,1:5]
The same procedure was used for phyla and KEGG pathways
Several dietary were found and eliminated before the rarefaction. We found contaminant phyla such as Chordata, Streptophyta, Arthropoda, etc.
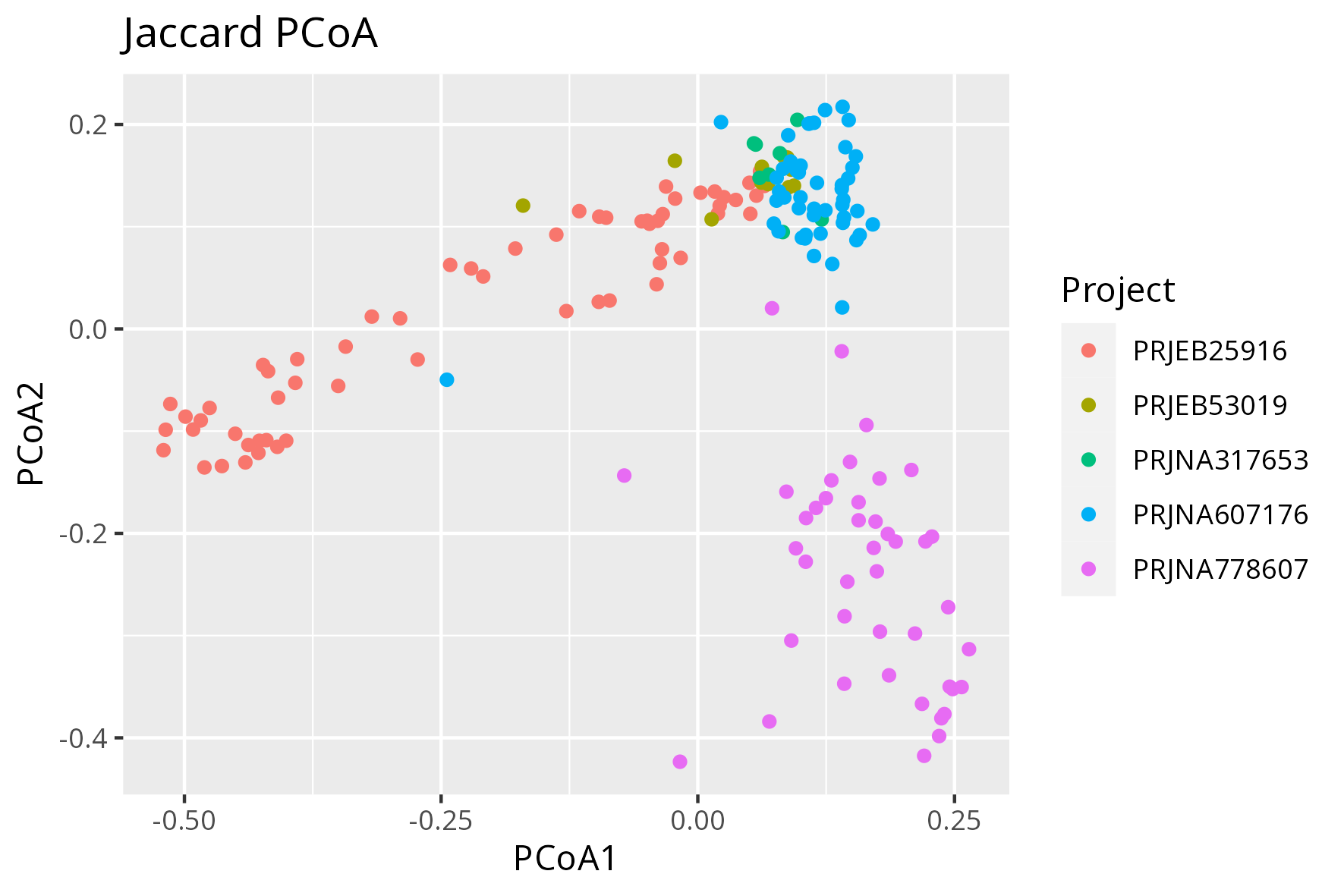 Figure A: Beta diversity - Jaccard PCoA representing all samples (171 healthy controls)
Figure A: Beta diversity - Jaccard PCoA representing all samples (171 healthy controls)
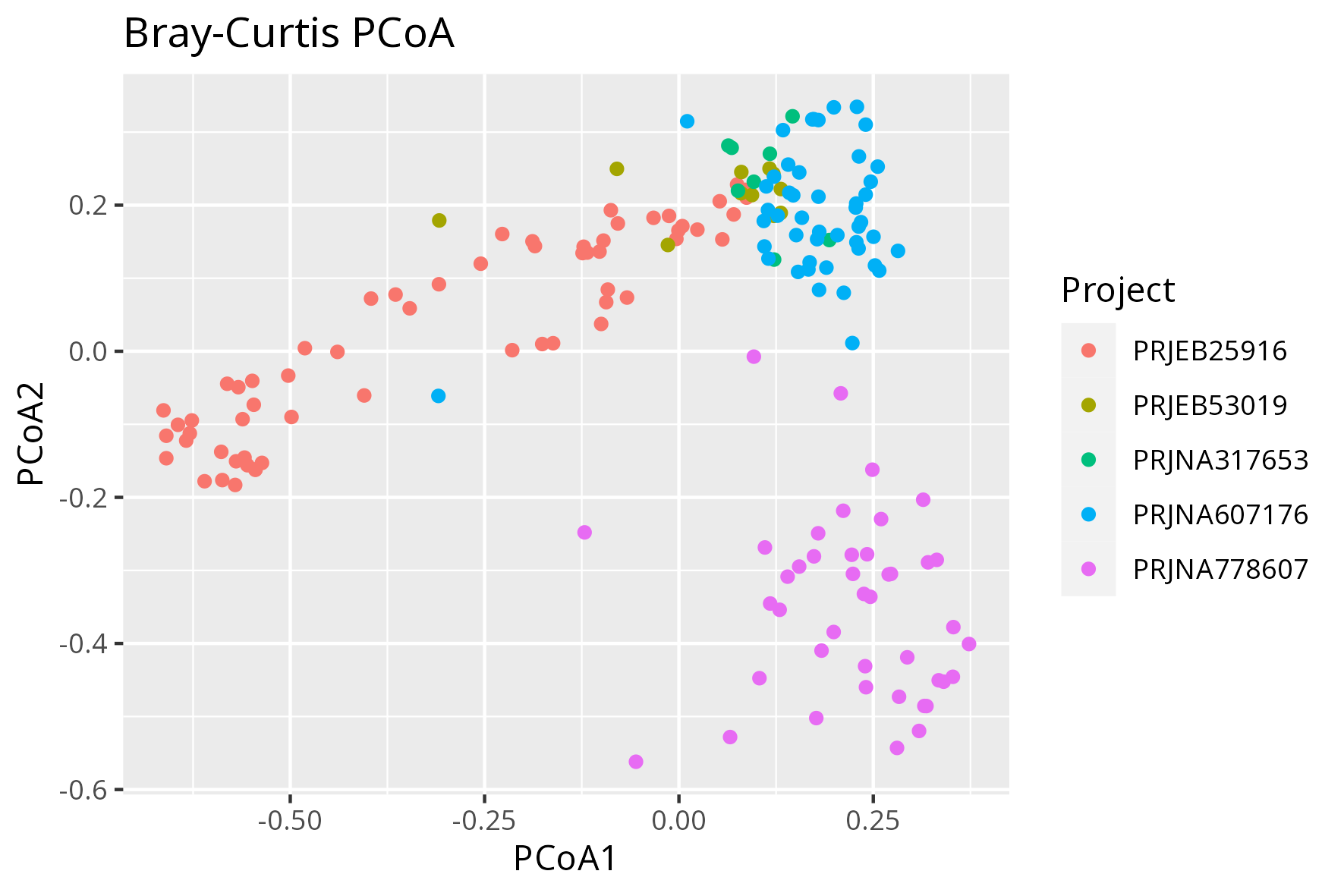 Figure B: Beta diversity - Bray-Curtis PCoA representing all samples (171 healthy controls)
Figure B: Beta diversity - Bray-Curtis PCoA representing all samples (171 healthy controls)
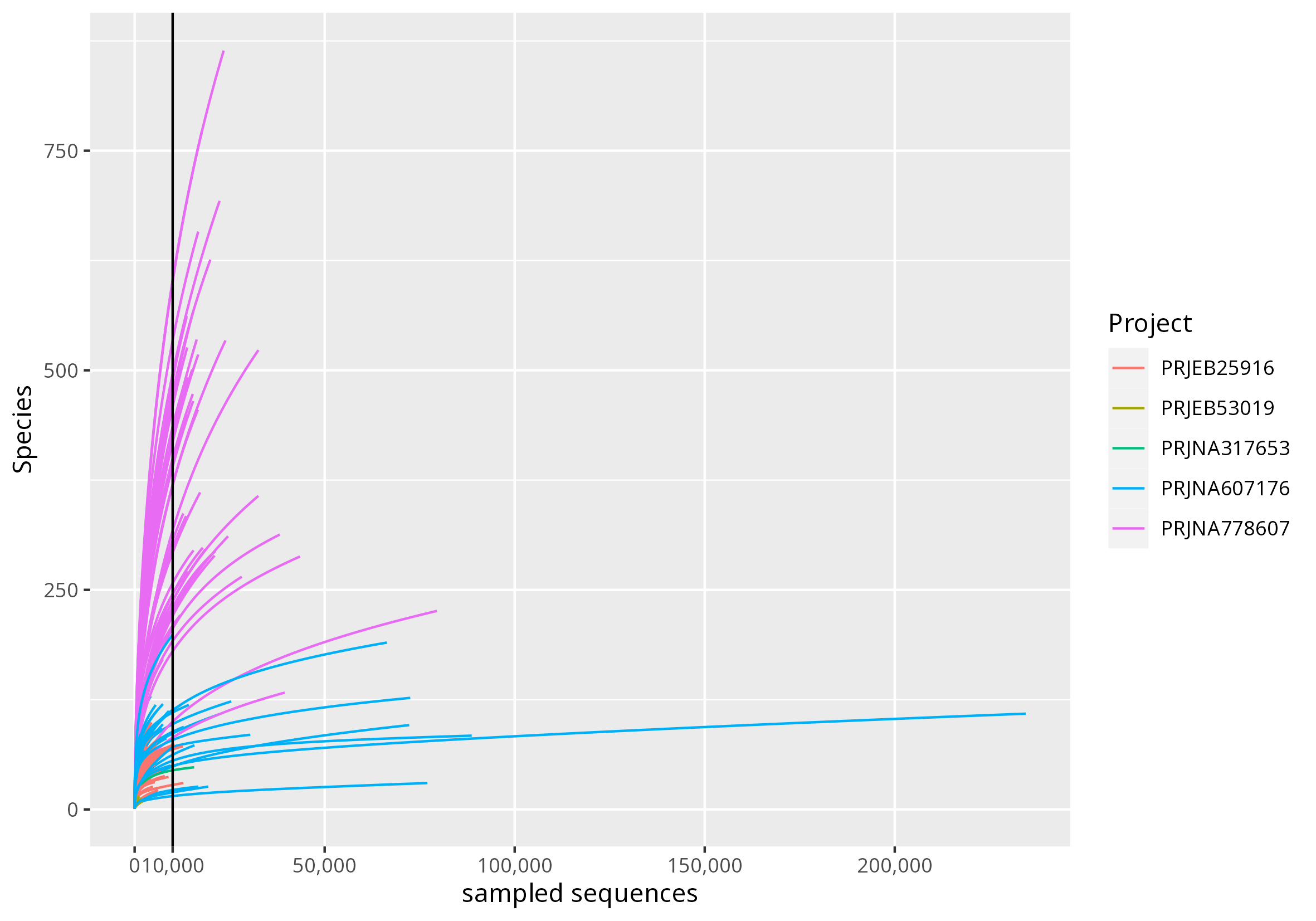 Figure C: Rarefaction curve. The selected sampling depth (10,000 reads) did not
guarantee to reach the plateau. It was selected in order to retain the largest
possible number of samples
Figure C: Rarefaction curve. The selected sampling depth (10,000 reads) did not
guarantee to reach the plateau. It was selected in order to retain the largest
possible number of samples
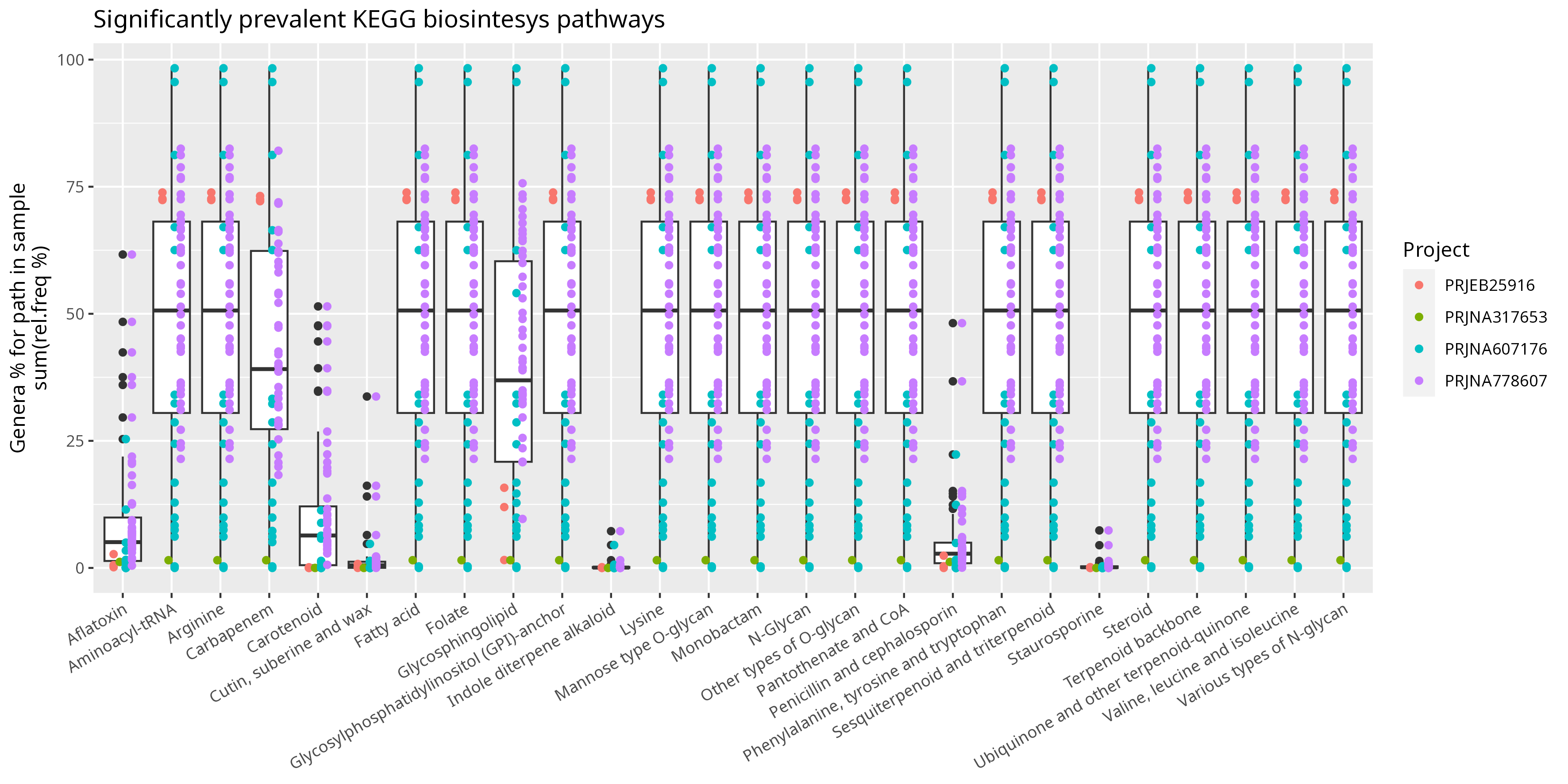 Figure D: Box-plot representing relative abundance sum of genera sustaining
KEGG prevalent biosynthesis pathways.
Figure D: Box-plot representing relative abundance sum of genera sustaining
KEGG prevalent biosynthesis pathways.
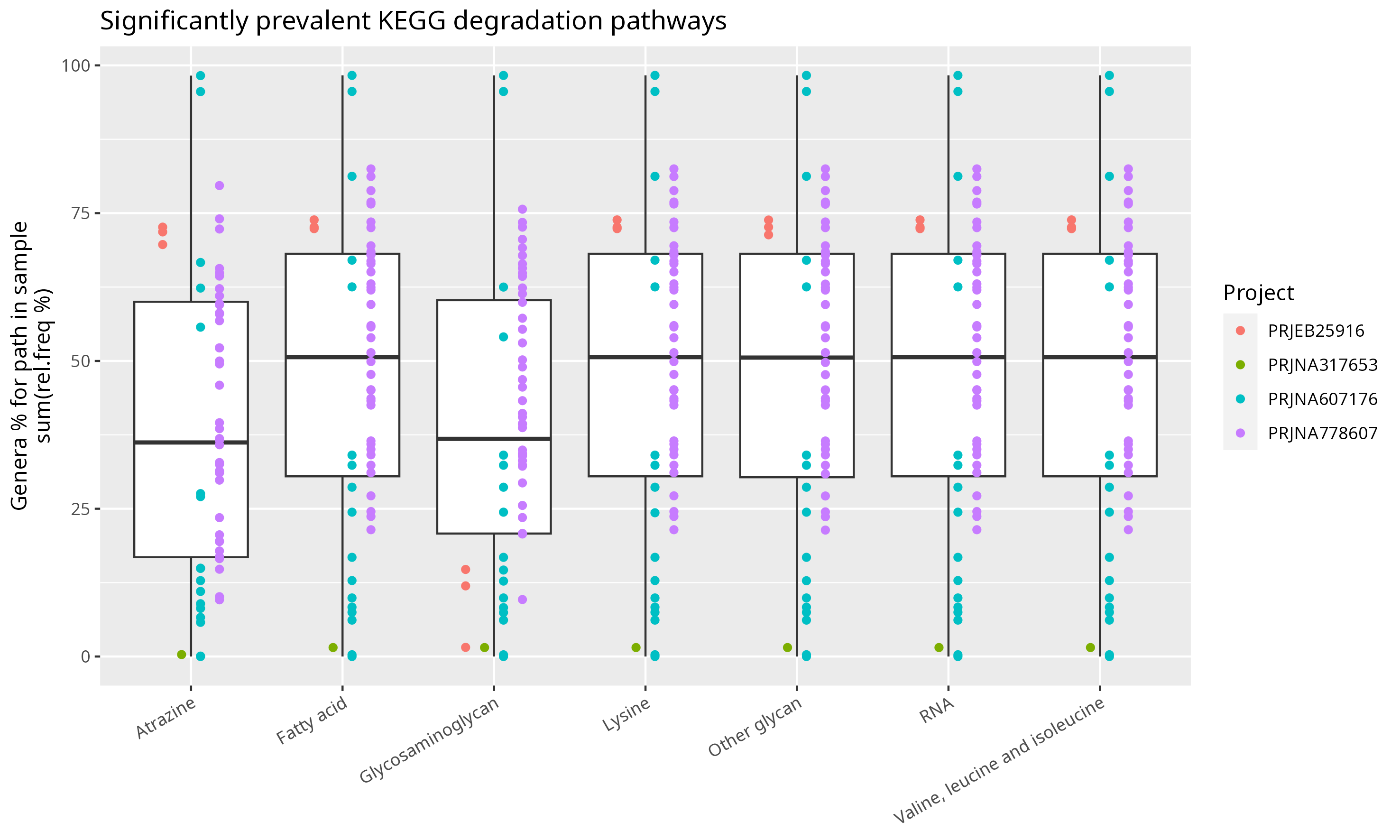 Figure E: Box-plot representing relative abundance sum of genera sustaining
KEGG prevalent degradation pathways.
Figure E: Box-plot representing relative abundance sum of genera sustaining
KEGG prevalent degradation pathways.
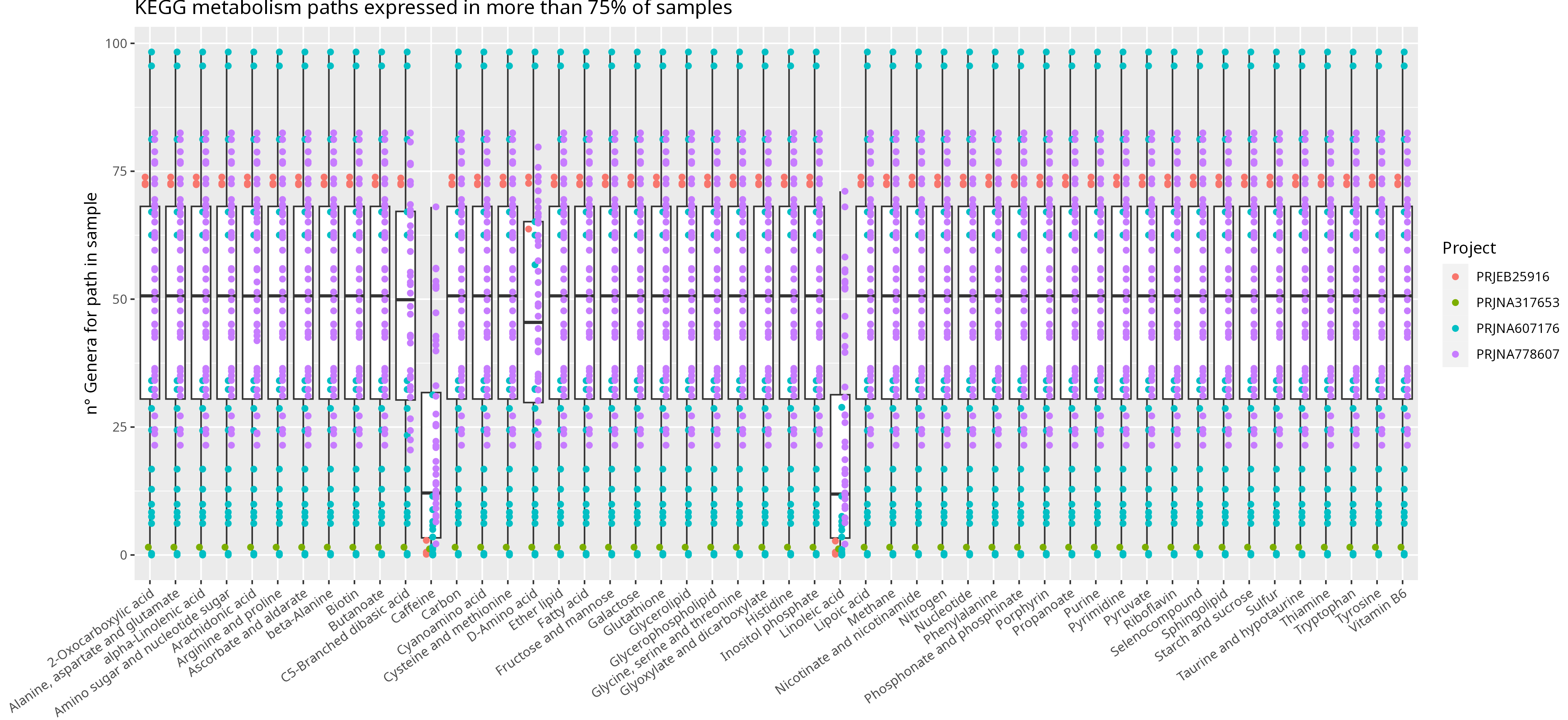 Figure D: Box-plot representing relative abundance sum of genera sustaining
KEGG prevalent metabolism pathways.
Figure D: Box-plot representing relative abundance sum of genera sustaining
KEGG prevalent metabolism pathways.
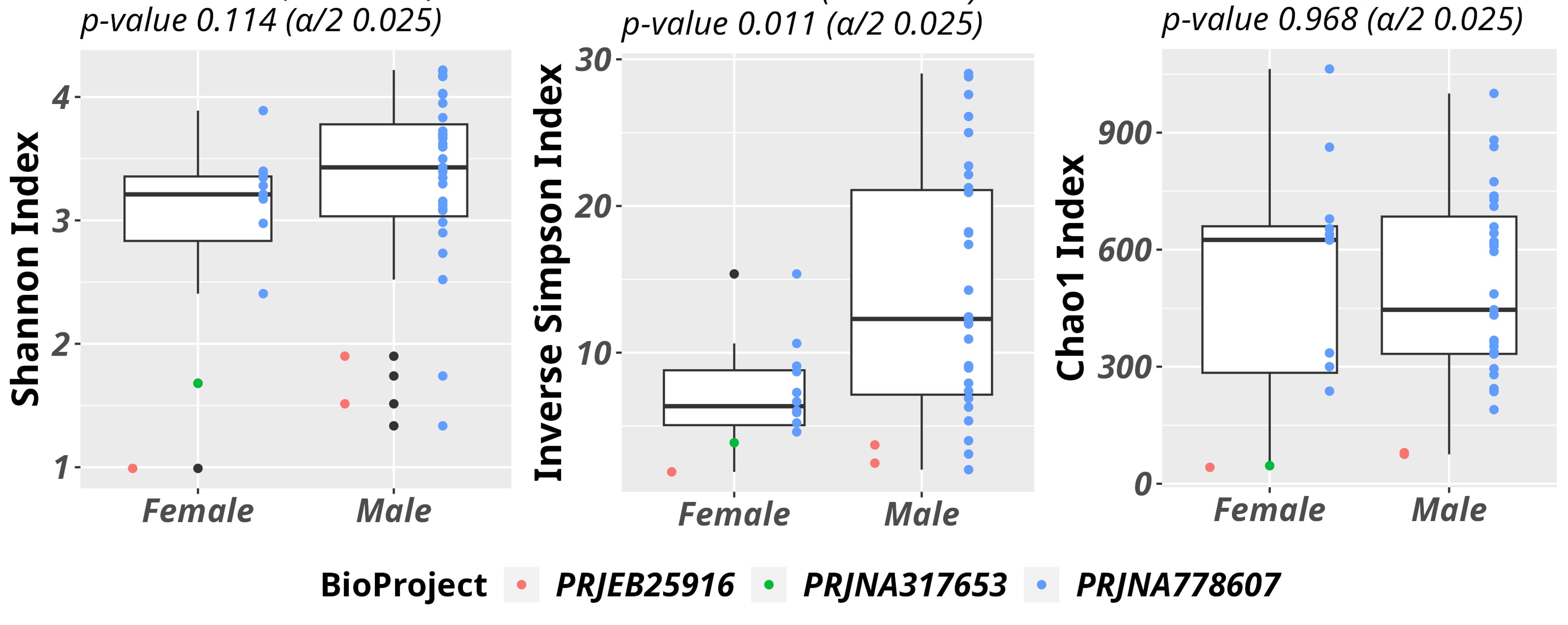 Figure D: Box plots representing Shannon, Inv. Simpson and Chao1 α-diversity indexes
for a group of samples for which gender metadata were available(12 Female vs 31 Male).
Wilcoxon tests were performed and resulting p-values are reported at the top of the box plots.
.
Figure D: Box plots representing Shannon, Inv. Simpson and Chao1 α-diversity indexes
for a group of samples for which gender metadata were available(12 Female vs 31 Male).
Wilcoxon tests were performed and resulting p-values are reported at the top of the box plots.
.
* The ITSoneWB web page is momentarily off. Then you can find the ITSoneWB docker
implementation at this link
and the BioMaS docker implementation.