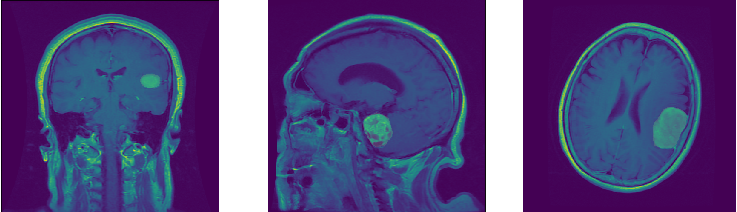
This repository contains implementation of the UNet[1], Deep ResUnet[2] Hybrid ResUnet and ONet[3] models. We tested those models` performance with the dice metric on the brain_tumor_dataset (https://figshare.com/articles/brain_tumor_dataset/1512427).
First download the code by git clone this repo:
git clone https://github.com/galprz/brain-tumor-segemntationThen use conda to install dependencies and setup the environment
conda end update -f environment.yml
conda activate brain-tumor-segmentationThe src folder contains all of the implementation code.
- utils/ contains helpers to download unzip and visualize the data.
- datasets.py file encapsulate the brain_tumor_dataset into pytorch datasets.
- loss.py and metrics.py contains the loss function and the dice evaluation metric correspondingly.
- models.py contains all the model implementation
- trainers.py contains the pytorch trainer implementation for the experiments.
The jupyter ipynb files contain all the experiments that we ran to evaluate the models` performance on our dataset. To reproduce the results we set the seed to constant value and the dataset will be downloaded automatically if you do not have the data folder so you can just rerun those files to reproduce the results.
- U-Net: Convolutional Networks for Biomedical Image Segmentation(https://arxiv.org/abs/1505.04597)
- Road Extraction by Deep Residual U-Net (https://arxiv.org/abs/1711.10684)
- Hybrid ResUnet and ONet (https://github.com/galprz/brain-tumor-segemntation/blob/master/report.pdf)