Website | NeurIPS 2021 Paper | Long Paper | Slack | TDC Mailing List |
Therapeutics Data Commons (TDC) is the first unifying framework to systematically access and evaluate machine learning across the entire range of therapeutics.
The collection of curated datasets, learning tasks, and benchmarks in TDC serves as a meeting point for domain and machine learning scientists. We envision that TDC can considerably accelerate machine-learning model development, validation and transition into biomedical and clinical implementation.
TDC is an open-source initiative. To get involved, check out the Contribution Guide!
Invited talk at the Harvard Symposium on Drugs for Future Pandemics (#futuretx20) [Slides] [Video]
Presented at NeurIPS 2021 [Poster] / Oral at ELLIS ML4Molecules [Slides] / Presented at Baylearn [Slides] [Poster]
The first TDC user group meeting will take place on Jan 25th 11AM-12:30PM EST! Find more info here!
0.3.4: Bug fixes on docking oracles, KL divergence measure, see commit 0f7121a and commit 6e46fbd!0.3.3: Extended support on cold split - now you can split based on multiple entities, see #127!0.3.2: Bug fixes - Adding support for harmonizing same DTIs with different affinities (KIBA, DAVIS Updated accordingly, see #98). Support label name retrieval for TWOSIDES (#121), and add gene symbol info to GDSC (#122).0.3.1: We have restructured the codebase to be contributor-friendly! Checkout the TDC documentation at https://tdc.readthedocs.io!- TDC paper is accepted to NeurIPS 2021 Datasets and Benchmarks
0.2.0: Release docking molecule generation benchmark! Checkout here!0.1.9: Support molecule filters! Checkout here!0.1.8: Streamlined and simplified the leaderboard programming frameworks! Now, you can submit a result for a single dataset! Checkout here!- TDC white paper is alive on arXiv!
Click here for older updates!
0.1.6: Released the second leaderboard on drug combination screening prediction! Checkout here!0.1.5: Added four realistic oracles from docking scores and synthetic accessibility! Checkout here!0.1.4: Added the 1st version ofMolConvertclass that can map among ~15 molecular formats in 2 lines of code (For 2D: from SMILES/SEFLIES and convert to SELFIES/SMILES, Graph2D, PyG, DGL, ECFP2-6, MACCS, Daylight, RDKit2D, Morgan, PubChem; For 3D: from XYZ, SDF files to Graph3D, Columb Matrix); Also a quality check on DTI datasets with IDs added.- Checkout Contribution Guide to add new dataset, task, function!
0.1.3: Added new therapeutics task on CRISPR Repair Outcome Prediction! Added a data function to map molecule to popular cheminformatics fingerprint.0.1.2: The first TDC Leaderboard is released! Checkout the leaderboard guide here and the ADMET Leaderboard here.0.1.1: Replaced VD, Half Life and Clearance datasets from new sources that have higher qualities. Added LD50 to Tox.0.1.0: Molecule quality check for ADME, Toxicity and HTS (canonicalized, and remove error mols).0.0.9: Added DrugComb NCI-60, CYP2C9/2D6/3A4 substrates, Carcinogens toxicity!0.0.8: Added hREG, DILI, Skin Reaction, Ames Mutagenicity, PPBR from AstraZeneca; added meta oracles!
- Diverse areas of therapeutics development: TDC covers a wide range of learning tasks, including target discovery, activity screening, efficacy, safety, and manufacturing across biomedical products, including small molecules, antibodies, and vaccines.
- Ready-to-use datasets: TDC is minimally dependent on external packages. Any TDC dataset can be retrieved using only 3 lines of code.
- Data functions: TDC provides extensive data functions, including data evaluators, meaningful data splits, data processors, and molecule generation oracles.
- Leaderboards: TDC provides benchmarks for fair model comparison and a systematic model development and evaluation.
- Open-source initiative: TDC is an open-source initiative. If you want to get involved, let us know.
To install the core environment dependencies of TDC, use pip:
pip install PyTDCNote: TDC is in the beta release. Please update your local copy regularly by
pip install PyTDC --upgradeThe core data loaders are lightweight with minimum dependency on external packages:
numpy, pandas, tqdm, scikit-learn, fuzzywuzzy, seabornFor utilities requiring extra dependencies, TDC prints installation instructions. To install full dependencies, please use the following conda-forge solution.
Data functions for molecule oracles, scaffold split, etc., require certain packages like RDKit. To install those packages, use the following conda installation:
conda install -c conda-forge pytdcWe provide many tutorials to get started with TDC:
| Name | Description |
|---|---|
| 101 | Introduce TDC Data Loaders |
| 102 | Introduce TDC Data Functions |
| 103.1 | Walk through TDC Small Molecule Datasets |
| 103.2 | Walk through TDC Biologics Datasets |
| 104 | Generate 21 ADME ML Predictors with 15 Lines of Code |
| 105 | Molecule Generation Oracles |
| 106 | Benchmark submission |
| DGL | Demo for DGL GNN User Group Meeting |
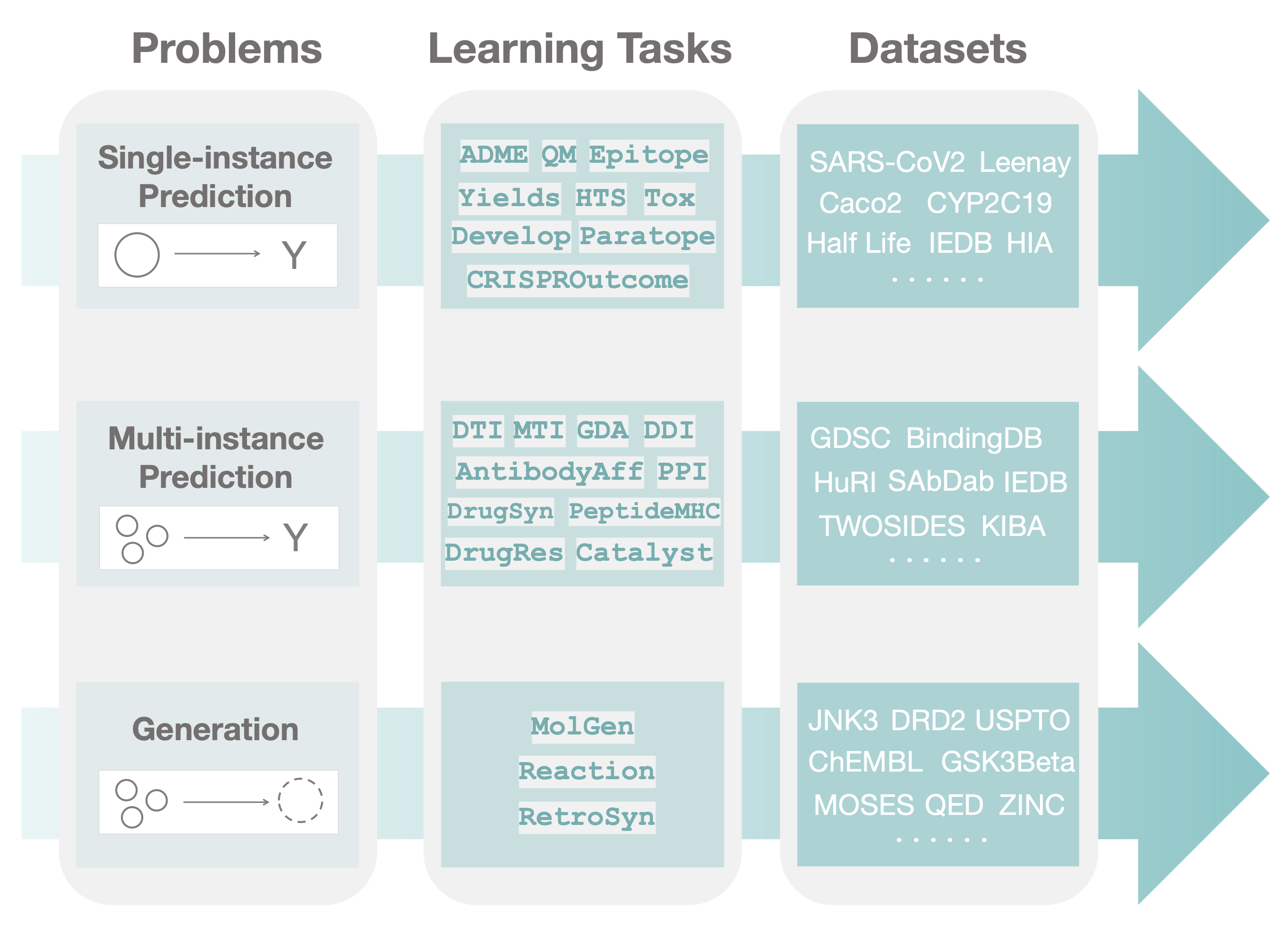
TDC has a unique three-tiered hierarchical structure, which to our knowledge, is the first attempt at systematically organizing machine learning for therapeutics. We organize TDC into three distinct problems. For each problem, we give a collection learning tasks. Finally, for each task, we provide a series of datasets.
In the first tier, after observing a large set of therapeutics tasks, we categorize and abstract out three major areas (i.e., problems) where machine learning can facilitate scientific advances, namely, single-instance prediction, multi-instance prediction, and generation:
- Single-instance prediction
single_pred: Prediction of property given individual biomedical entity. - Multi-instance prediction
multi_pred: Prediction of property given multiple biomedical entities. - Generation
generation: Generation of new desirable biomedical entities.
The second tier in the TDC structure is organized into learning tasks. Improvement on these tasks can result in numerous applications, including identifying personalized combinatorial therapies, designing novel class of antibodies, improving disease diagnosis, and finding new cures for emerging diseases.
Finally, in the third tier of TDC, each task is instantiated via multiple datasets. For each dataset, we provide several splits of the dataset into training, validation, and test sets to simulate the type of understanding and generalization (e.g., the model's ability to generalize to entirely unseen compounds or to granularly resolve patient response to a polytherapy) needed for transition into production and clinical implementation.
TDC provides a collection of workflows with intuitive, high-level APIs for both beginners and experts to create machine learning models in Python. Building off the modularized "Problem -- Learning Task -- Data Set" structure (see above) in TDC, we provide a three-layer API to access any learning task and dataset. This hierarchical API design allows us to easily incorporate new tasks and datasets.
For a concrete example, to obtain the HIA dataset from ADME therapeutic learning task in the single-instance prediction problem:
from tdc.single_pred import ADME
data = ADME(name = 'HIA_Hou')
# split into train/val/test with scaffold split methods
split = data.get_split(method = 'scaffold')
# get the entire data in the various formats
data.get_data(format = 'df')You can see all the datasets that belong to a task as follows:
from tdc.utils import retrieve_dataset_names
retrieve_dataset_names('ADME')See all therapeutic tasks and datasets on the TDC website!
To retrieve the training/validation/test dataset split, you could simply type
data = X(name = Y)
data.get_split(seed = 42)
# {'train': df_train, 'val': df_val, 'test': df_test}You can specify the splitting method, random seed, and split fractions in the function by e.g. data.get_split(method = 'scaffold', seed = 1, frac = [0.7, 0.1, 0.2]). Check out the data split page on the website for details.
We provide various evaluation metrics for the tasks in TDC, which are described in model evaluation page on the website. For example, to use metric ROC-AUC, you could simply type
from tdc import Evaluator
evaluator = Evaluator(name = 'ROC-AUC')
score = evaluator(y_true, y_pred)TDC provides numerous data processing functions, including label transformation, data balancing, pair data to PyG/DGL graphs, negative sampling, database querying and so on. For function usage, see our data processing page on the TDC website.
For molecule generation tasks, we provide 10+ oracles for both goal-oriented and distribution learning. For detailed usage of each oracle, please checkout the oracle page on the website. For example, we want to retrieve the GSK3Beta oracle:
from tdc import Oracle
oracle = Oracle(name = 'GSK3B')
oracle(['CC(C)(C)....'
'C[C@@H]1....',
'CCNC(=O)....',
'C[C@@H]1....'])
# [0.03, 0.02, 0.0, 0.1]Every dataset in TDC is a benchmark, and we provide training/validation and test sets for it, together with data splits and performance evaluation metrics. To participate in the leaderboard for a specific benchmark, follow these steps:
-
Use the TDC benchmark data loader to retrieve the benchmark.
-
Use training and/or validation set to train your model.
-
Use the TDC model evaluator to calculate the performance of your model on the test set.
-
Submit the test set performance to a TDC leaderboard.
As many datasets share a therapeutics theme, we organize benchmarks into meaningfully defined groups, which we refer to as benchmark groups. Datasets and tasks within a benchmark group are carefully curated and centered around a theme (for example, TDC contains a benchmark group to support ML predictions of the ADMET properties). While every benchmark group consists of multiple benchmarks, it is possible to separately submit results for each benchmark in the group. Here is the code framework to access the benchmarks:
from tdc import BenchmarkGroup
group = BenchmarkGroup(name = 'ADMET_Group', path = 'data/')
predictions_list = []
for seed in [1, 2, 3, 4, 5]:
benchmark = group.get('Caco2_Wang')
# all benchmark names in a benchmark group are stored in group.dataset_names
predictions = {}
name = benchmark['name']
train_val, test = benchmark['train_val'], benchmark['test']
train, valid = group.get_train_valid_split(benchmark = name, split_type = 'default', seed = seed)
# --------------------------------------------- #
# Train your model using train, valid, test #
# Save test prediction in y_pred_test variable #
# --------------------------------------------- #
predictions[name] = y_pred_test
predictions_list.append(predictions)
results = group.evaluate_many(predictions_list)
# {'caco2_wang': [6.328, 0.101]}For more information, please visit here.
If you find Therapeutics Data Commons useful, consider citing our NeurIPS paper:
@article{Huang2021tdc,
title={Therapeutics Data Commons: Machine Learning Datasets and Tasks for Drug Discovery and Development},
author={Huang, Kexin and Fu, Tianfan and Gao, Wenhao and Zhao, Yue and Roohani, Yusuf and Leskovec, Jure and Coley,
Connor W and Xiao, Cao and Sun, Jimeng and Zitnik, Marinka},
journal={Proceedings of Neural Information Processing Systems, NeurIPS Datasets and Benchmarks},
year={2021}
}
TDC is a community-driven and open-science initiative. If you want to get involved, join the Slack Workspace and checkout the contribution guide!
Send emails to us or open an issue.
TDC is hosted on Harvard Dataverse with the following persistent identifier https://doi.org/10.7910/DVN/21LKWG. When Dataverse is under maintenance, TDC datasets cannot be retrieved. That happens rarely; please check the status on the Dataverse website.
TDC codebase is under MIT license. For individual dataset usage, please refer to the dataset license found in the website.