cleanRfield: A tool for cleaning and filtering data spatial points from crop yield maps in R.
This package is a compilation of functions to clean and filter observations from yield monitors or other agricultural spatial point data. Yield monitors are prone to error, and filtering the observations or removing observations from near field boundaries can improve estimates of whole-field yield, combine speed, grain moisture, or other parameters of interest. In this package, users can easily select filters for one or more traits and prepare a smaller dataset to make decisions.
This tutorial assumes that readers have a basic understanding of spatial data, including projections and coordinate reference systems. If you need a refresher on this topic, we recommend reading this blog post for deciding between projected and unprojected data and this post for understanding the basics of coordinate reference systems.
2. Cropping or selecting regions
6. Buffering the field boundaries
7. Filtering using data values
8. Filtering using standard deviation values
9. Evaluating multiple fields on parallel
12. Working with .csv or .txt files
Now install R/cleanRfield using the
install_github()function from devtools package. If necessary, use the argument type="source".
install.packages("devtools")
devtools::install_github("filipematias23/cleanRfield")Calling necessary packages:
library(terra)
library(cleanRfield)Start this tutorial by downloading the example EX1 here and the field boundary here. This tutorial will use the function
vect()from package terra to read and upload the data to RStudio (see provided code).
EX1 is a yield map from a soybean field, stored as a point shapefile. Yield monitor observations were originally collected in the north-central US using a combine yield monitor, and observations were geographically shifted to protect the landowner’s privacy. This data set include three attributes:
- Speed (miles per hour): speed of the combine at the time the observation was recorded
- Dry_Yield (bushels per acre): yield at that observation’s location as recorded by the yield monitor, adjusted to a 13% moisture basis
- Adj_Moist (percent): indicates what moisture level the original wet yield measurements were adjusted to when calculating dry yield
The field boundary is a shapefile layer with three (3) polygons that delineate the boundaries of the sample field EX1.
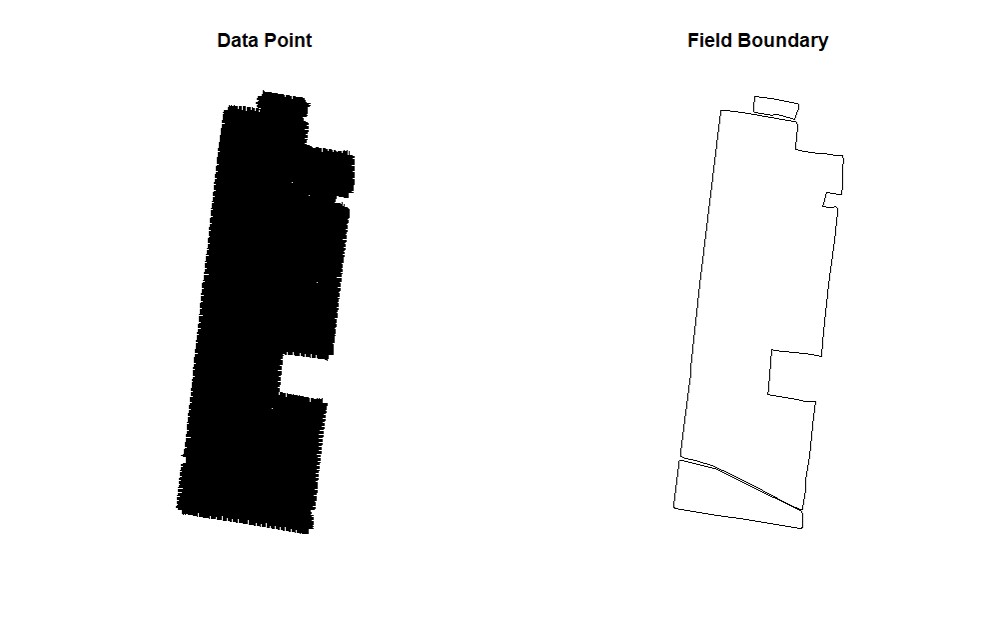
### Opening Sample Field 1 ###
par(mfrow=c(1,2))
EX1<-vect("EX1.shp")
plot(EX1, main="Data Point")
EX1.Shape<-vect("EX1_boundary.shp")
plot(EX1.Shape, main="Field Boundary")
par(mfrow=c(1,1))Users can subset the data by drawing boundaries around a field or subset of fields. Function
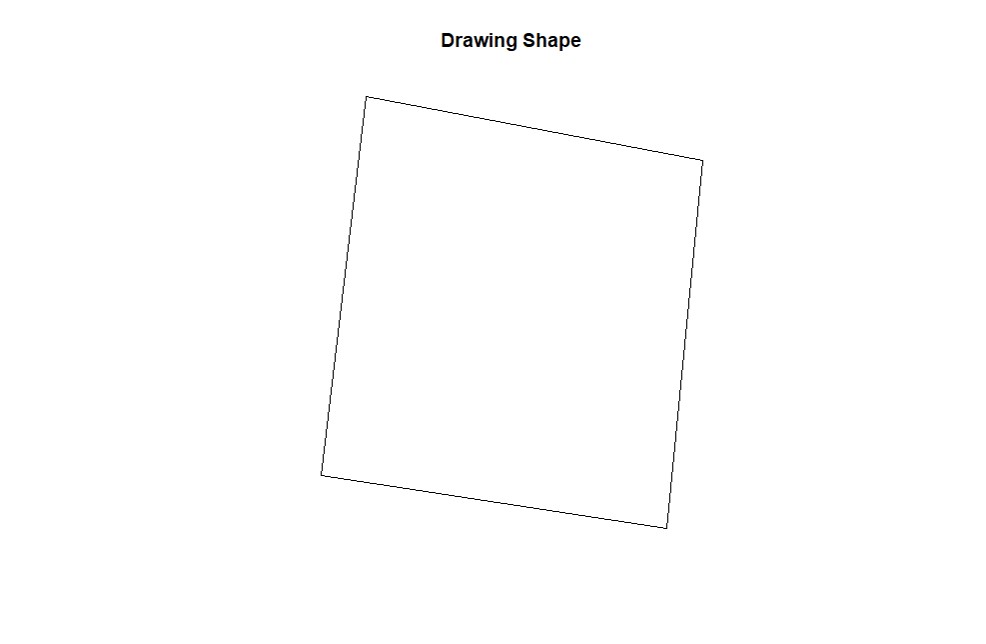
cropField. Depending on your computer and the size of your data set, this step may take a few seconds. This function works better using a popup window, not RStudio's integrated viewing pane, so we've included code for opening that new window.
#Open an extra plot window
x11()
# "Use cursor to select 4 points around polygon (1) in the plots window."
EX1.C<-cropField(field = EX1, nPolygon = 1, nPoint = 4)plot(EX1.C$shape,main="Drawing Shape")# Using the shape drawn above to crop data:
EX1.C<-cropField(field = EX1, shape = EX1.C$shape) # All data will be selected using the full boundary as shape:
EX1.C1<-cropField(field = EX1, shape = EX1.Shape) The newest version of RStudio (2021.09.2) has updated the plot viewing pane. If you are using the newest RStudio, you may need run an additional line of code to open the point-and-click cropping functionality in a pop-out window instead of the integrated plot viewing pane.
#Open an extra plot window
x11()
# "Use cursor to select 4 points around polygon (1) in the plots window."
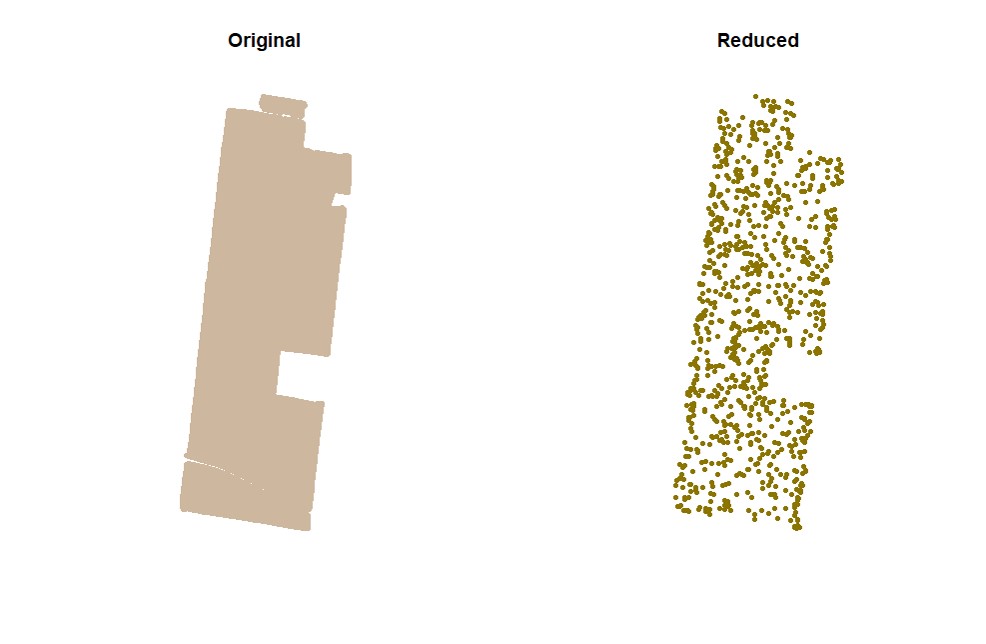
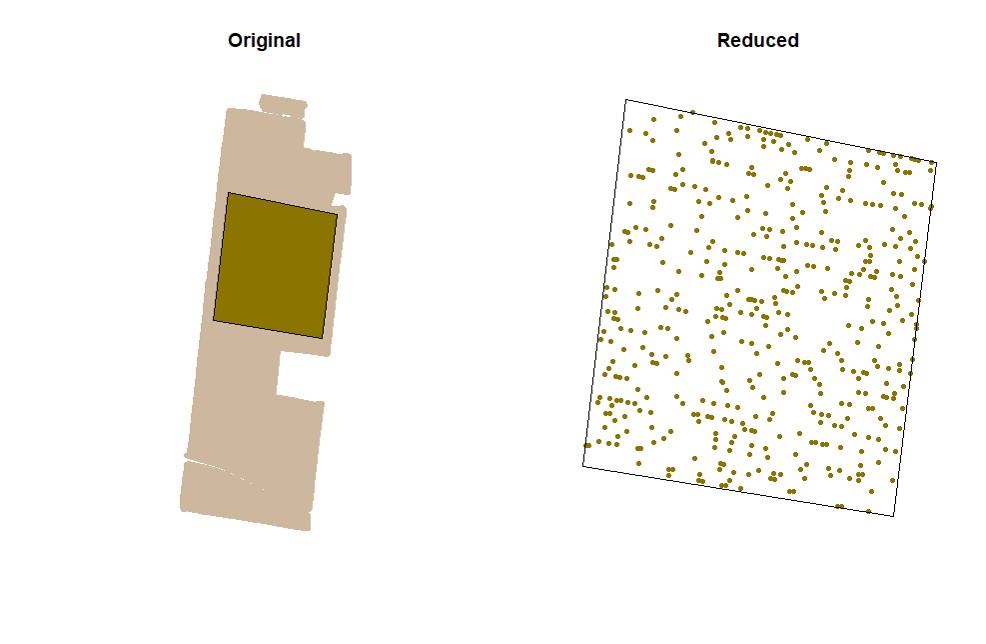
EX1.C<-cropField(field = EX1, nPolygon = 1, nPoint = 4)Users can sample random points in the data. Function
sampleField.
# Sampling 5%:
EX1.S<-sampleField(field = EX1, size = 0.05) # Sampling 10% under a small shape:
EX1.S<-sampleField(field = EX1,shape = EX1.C$shape, size = 0.1) # Sampling 10% under a full shape:
EX1.S<-sampleField(field = EX1,shape = EX1.Shape, size = 0.1) Function
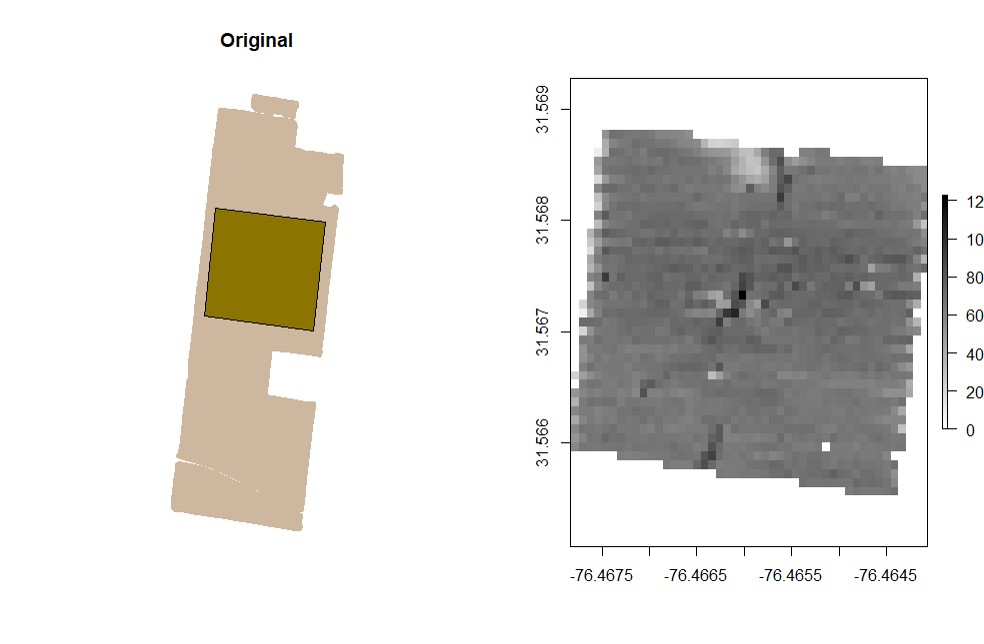
rasterField.Data points can be used to create raster files. Use either the provided code for unprojected data or projected data-- you will not need to run both sets of code. Any positive number can be chosen as the resolution, but choosing too high of a resolution will result in a raster file that oversimplifies the shape of the field, and choosing too low of a resolution can cause the runtime to be to long and/or cause the parts of the field between combine passes to be excluded from the final field shape.
# Check projection to observe 'LENGTHUNIT':
crs(EX1)
# Unprojected Data (non or NA): use resolution around 0.00008 to create a raster for "Dry_Yield":
EX1.R<-rasterField(field = EX1,
trait = c("Dry_Yield"),
res = 0.00008)
# Projected Data (e.g., +units=m or +units=us-ft): use resolution around 5 to 20 to create a raster for "Dry_Yield":
EX1.R<-rasterField(field = EX1,
trait = c("Dry_Yield"),
res = 20) # Making raster only for the small shape:
EX1.R<-rasterField(field = EX1,
shape = EX1.C$shape,
trait = c("Dry_Yield"),
res = 0.00008) # Attention: for projected data use res=20 (e.g., +units=m or +units=us-ft).# Multilayer raster for two or more traits:
EX1.R<-rasterField(field = EX1,
trait = c("Dry_Yield","Speed"),
res = 0.00008) # Attention: for projected data use res=20 (e.g., +units=m or +units=us-ft).# Different raster color visualizations:
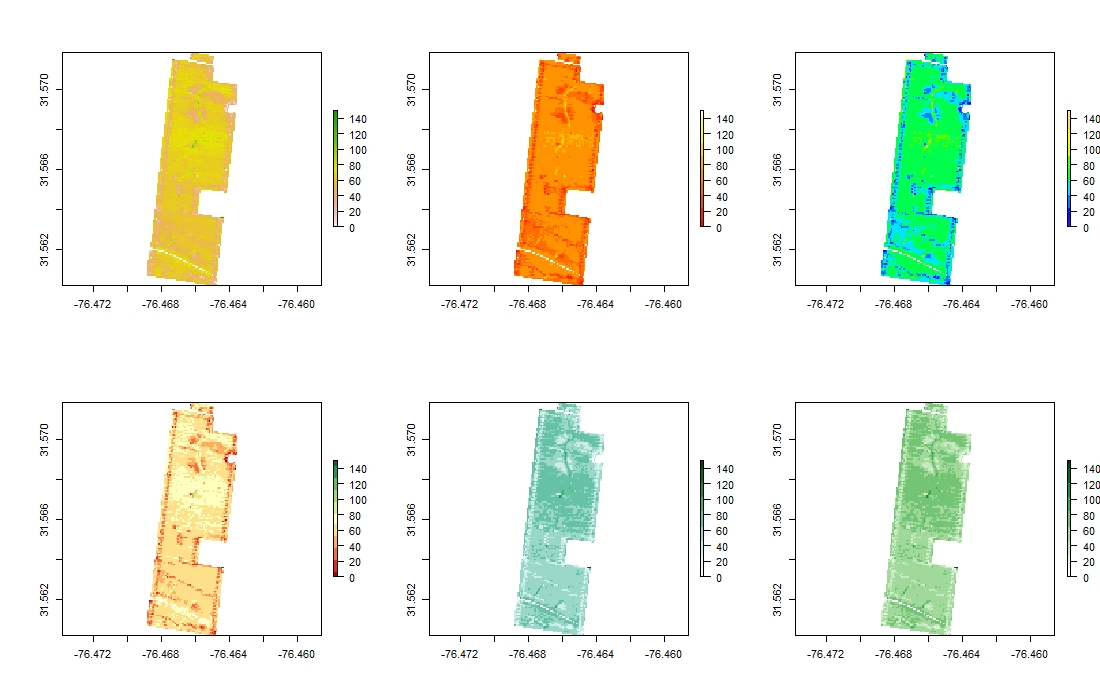
library(RColorBrewer)
par(mfrow=c(2,3))
plot(EX1.R$Dry_Yield)
plot(EX1.R$Dry_Yield,col = heat.colors(10))
plot(EX1.R$Dry_Yield,col = topo.colors(10))
plot(EX1.R$Dry_Yield,col = brewer.pal(11, "RdYlGn"))
plot(EX1.R$Dry_Yield,col = brewer.pal(9, "BuGn"))
plot(EX1.R$Dry_Yield,col = brewer.pal(9, "Greens"))
par(mfrow=c(1,1))Users can manually draw field boundaries or use the raster layer to draw field boundaries automatically. Function
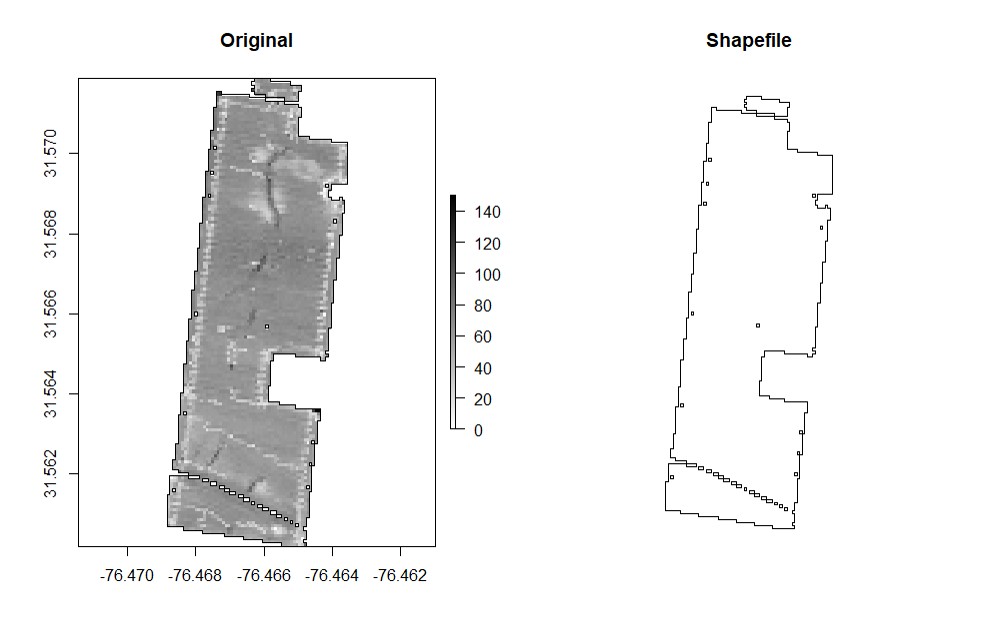
boundaryField.
- Automatic - a raster layer is necessary for drawing the boundary automatically, which is the fastest method (use function
rasterFieldbefore). Increasing the tolerance parameter simplifies the geometry of complex boundaries.
EX1.P<-boundaryField(field = EX1.R$Dry_Yield, tolerance = 0.0004) # Yield data did not capture borders of field
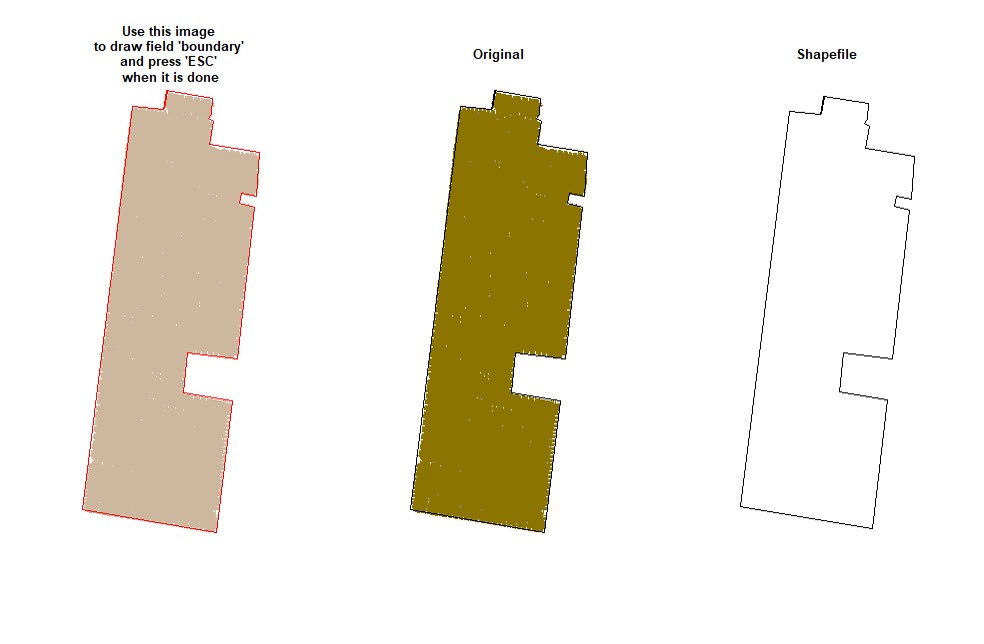
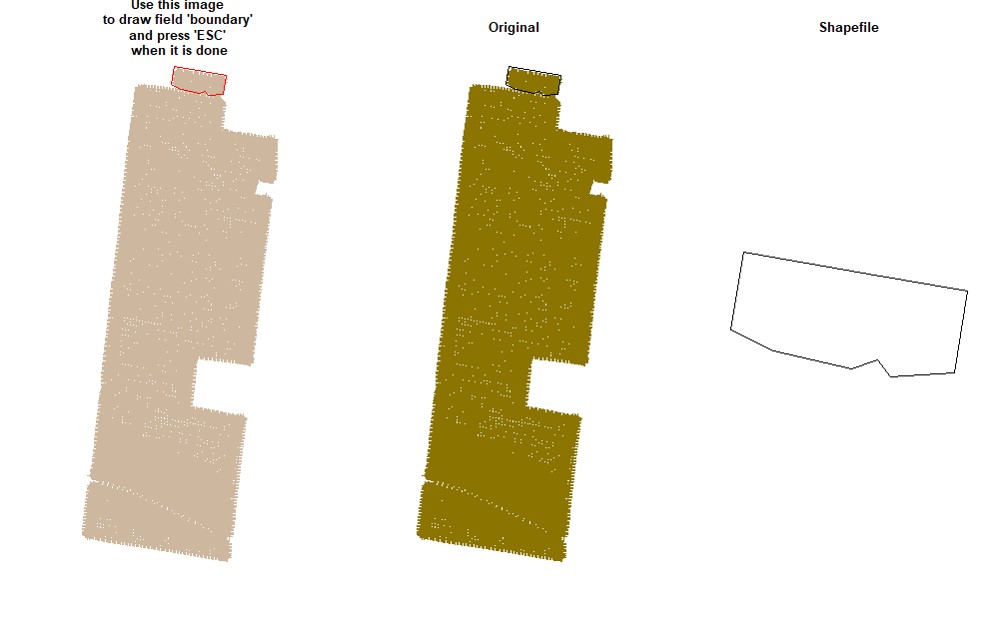
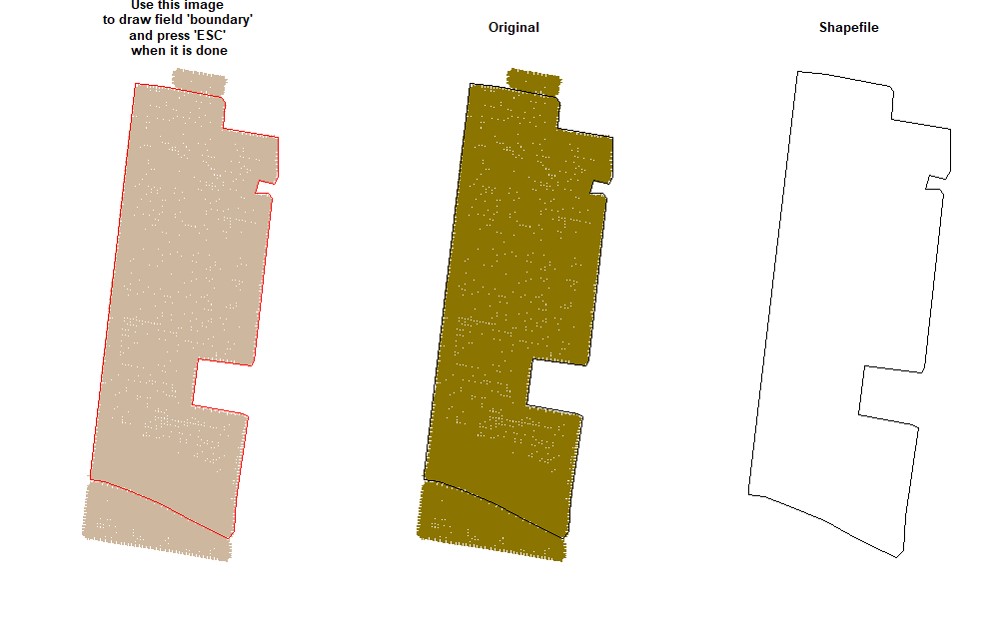
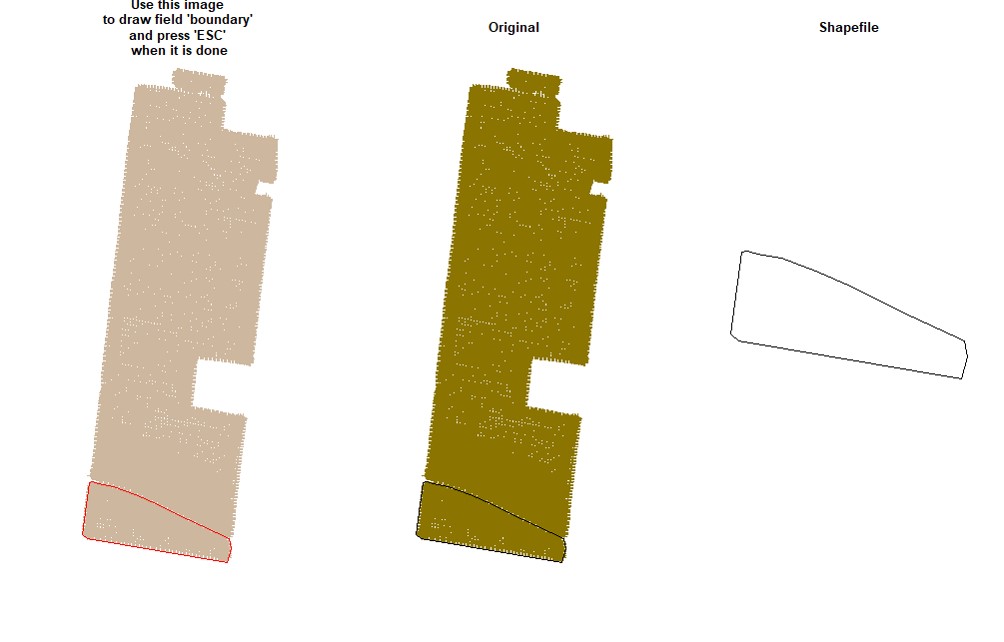
EX1.P<-boundaryField(field = EX1.R$Speed) # Speed data has defined field borders, we use default tolerance- Manually - use your cursor to make points around the field boundary and press ESC when it is done (use the parameter
draw = TRUE).
EX1.P<-boundaryField(field = EX1, draw = TRUE)- Drawing 3 different fields (Manually):
# Upper field:
EX1.P1<-boundaryField(field = EX1, draw = T) # Manually# Middle field:
EX1.P2<-boundaryField(field = EX1, draw = T)# Lower field:
EX1.P3<-boundaryField(field = EX1, draw = T)- Combining fields on the same shapefile:
# Combining field on the same shapefile:
EX1.P<-rbind(EX1.P1,EX1.P2,EX1.P3)
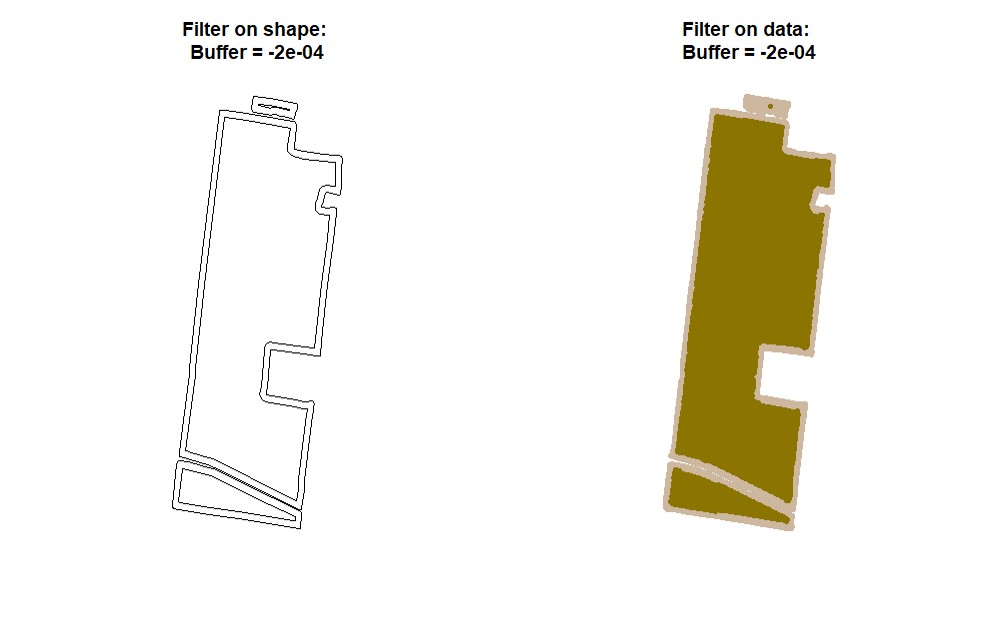
plot(EX1.P)Users can make a buffer around the field boundarys using a new shapefile (Value must be negative). Function
bufferField. Like in section 4, use either the provided code for unprojected data or projected data-- you will not need to run both sets of code. Under the cleanRfield update from March 2023, most input data sets will now be treated as projected data, even if they have the CRS WGS84. Be sure to check the 'LENGTHUNIT' atribute.
- Only shapefile:
# Check projection to observe 'LENGTHUNIT':
crs(EX1)
# Unprojected Data (e.g., non or NA): buffer of -0.0001:
EX1.B<-bufferField(shape = EX1.Shape,value = -0.0001)
# Projected Data (e.g., LENGTHUNIT = metre, +units=us-ft, or other linear units): buffer of -5:
EX1.B<-bufferField(shape = EX1.Shape, value = -5)
- Making buffering and automatic filtering at the same time (Shapefile + Data).
# Buffer of -15 metres:
EX1.B<-bufferField(shape = EX1.Shape,
field = EX1,
value = -15) # Attention: check your units before assigning "value."Users can filter spatial point data using values criteria. Function
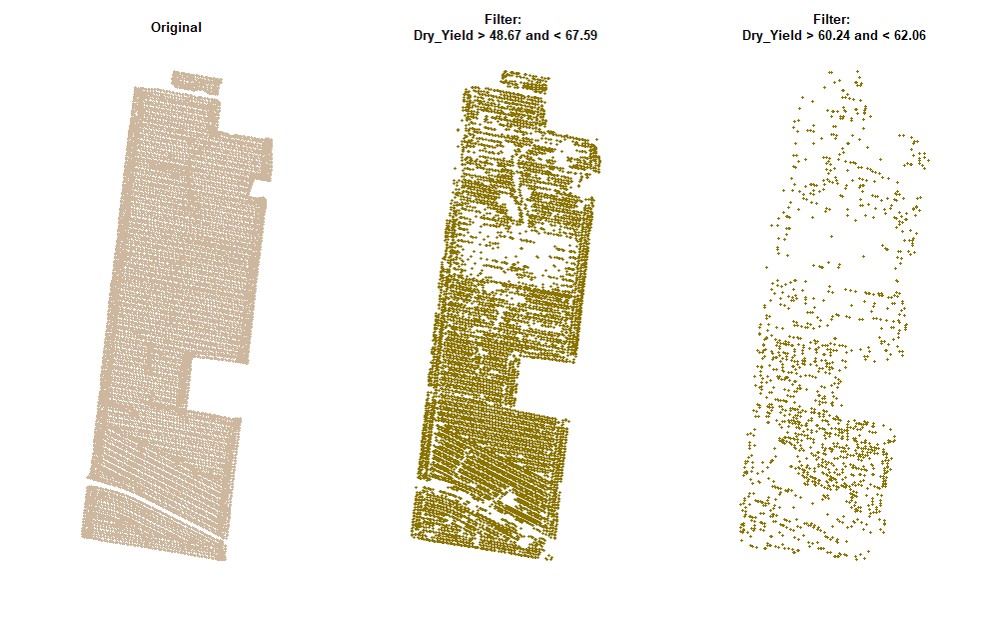
filterField.
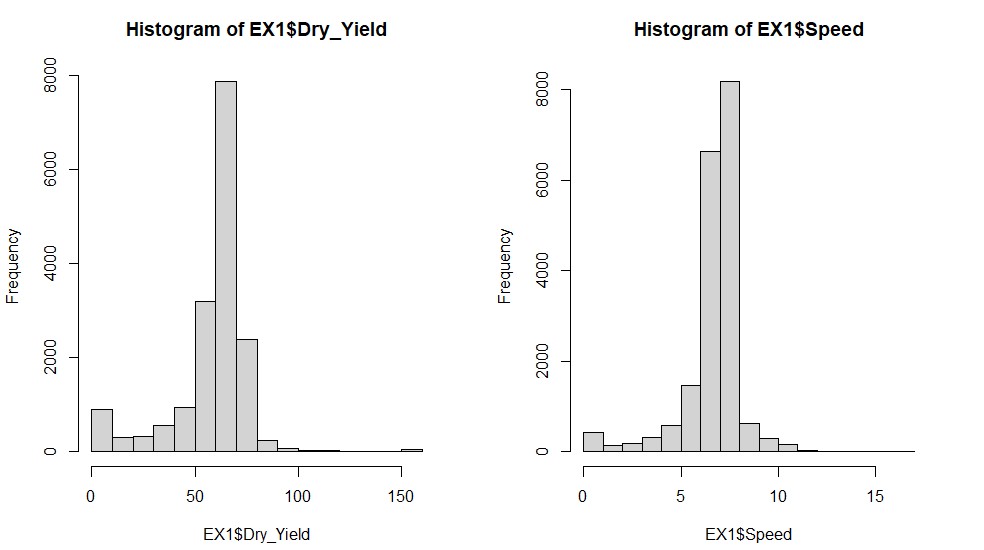
- Observing traits histograms:
par(mfrow=c(1,2))
hist(EX1$Dry_Yield)
hist(EX1$Speed)
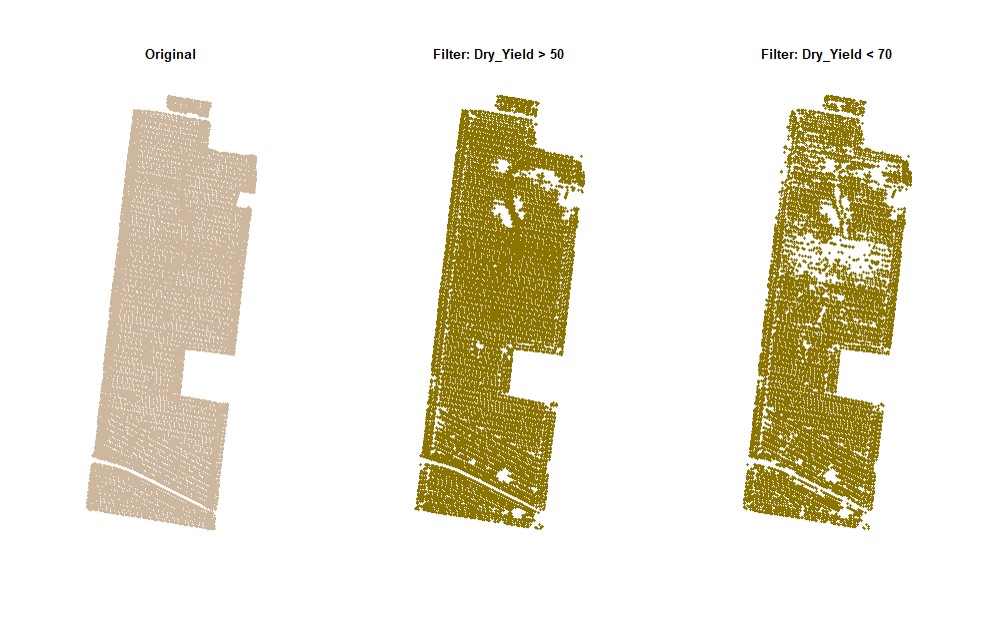
par(mfrow=c(1,1))# Filtering data for Dry_Yield>60:
EX1.F<-filterField(field = EX1,
trait = "Dry_Yield",
value = 60,
cropAbove = T)# Filtering data for Dry_Yield>50 and Dry_Yield<70:
EX1.F<-filterField(field = EX1,
trait = c("Dry_Yield","Dry_Yield"),
value = c(50,70),
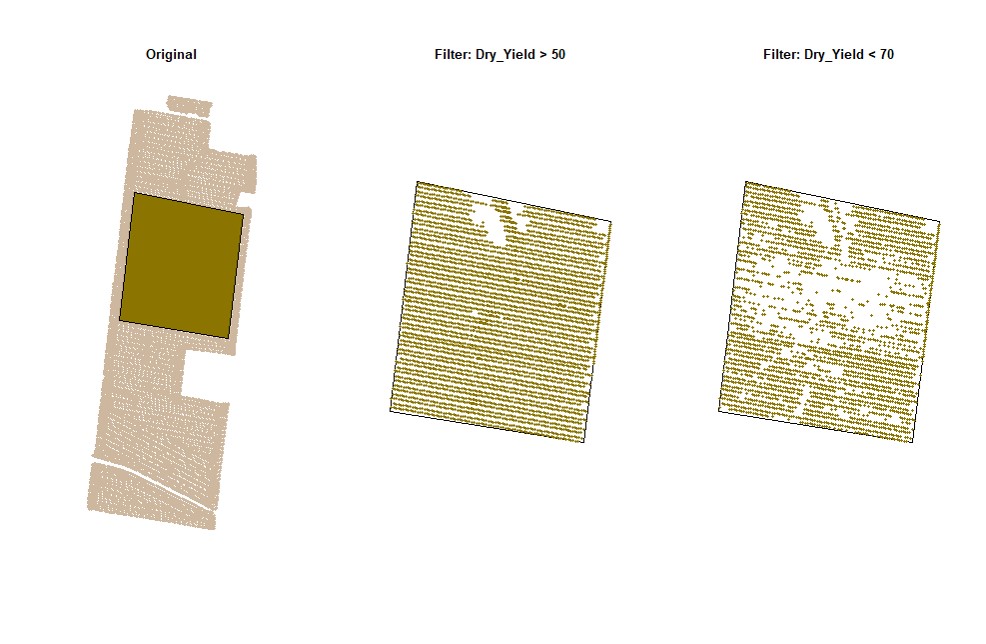
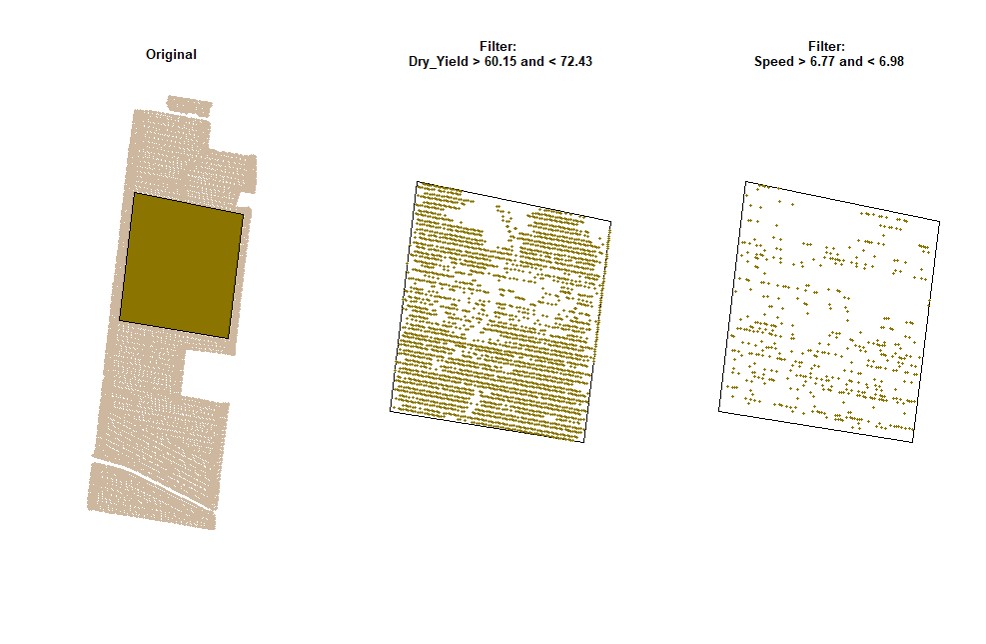
cropAbove = c(T,F))# Filtering data for Dry_Yield>50 and Dry_Yield<70 (only for the data on the small shapefile):
EX1.F<-filterField(field = EX1,
shape = EX1.C$shape,
trait = c("Dry_Yield","Dry_Yield"),
value = c(50,70),
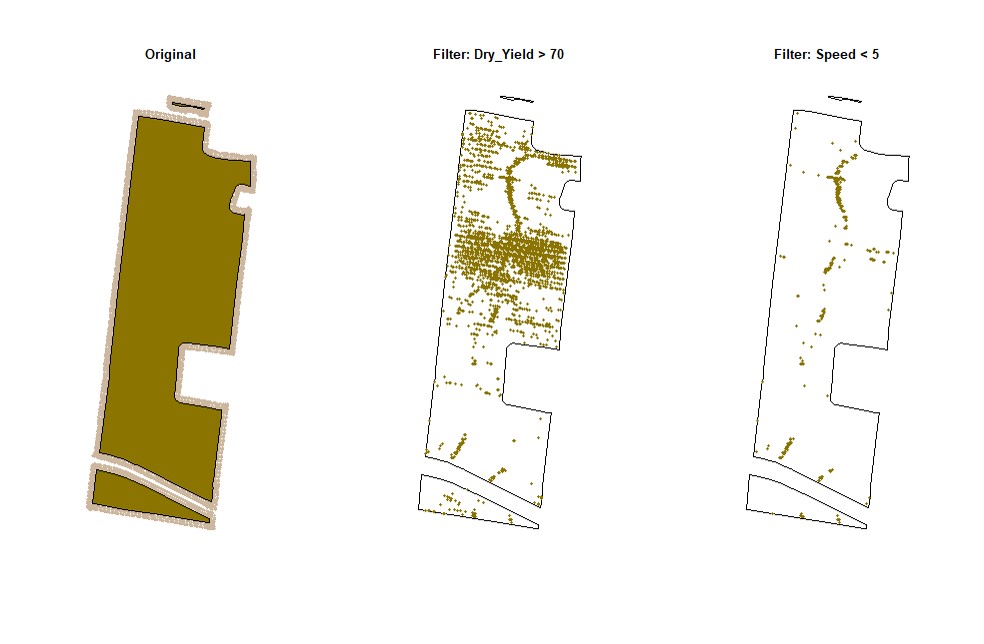
cropAbove = c(T,F))# Filtering data for Dry_Yield>70 and Speed<5 (using the buffer shapefile):
EX1.F<-filterField(field = EX1,
shape = EX1.B$newShape,
trait = c("Dry_Yield","Speed"),
value = c(70,5),
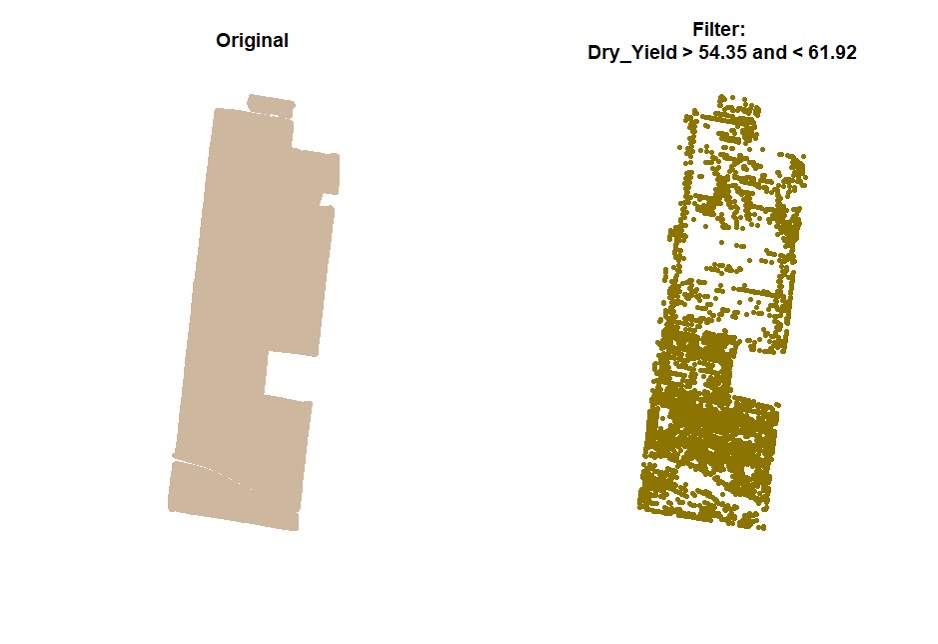
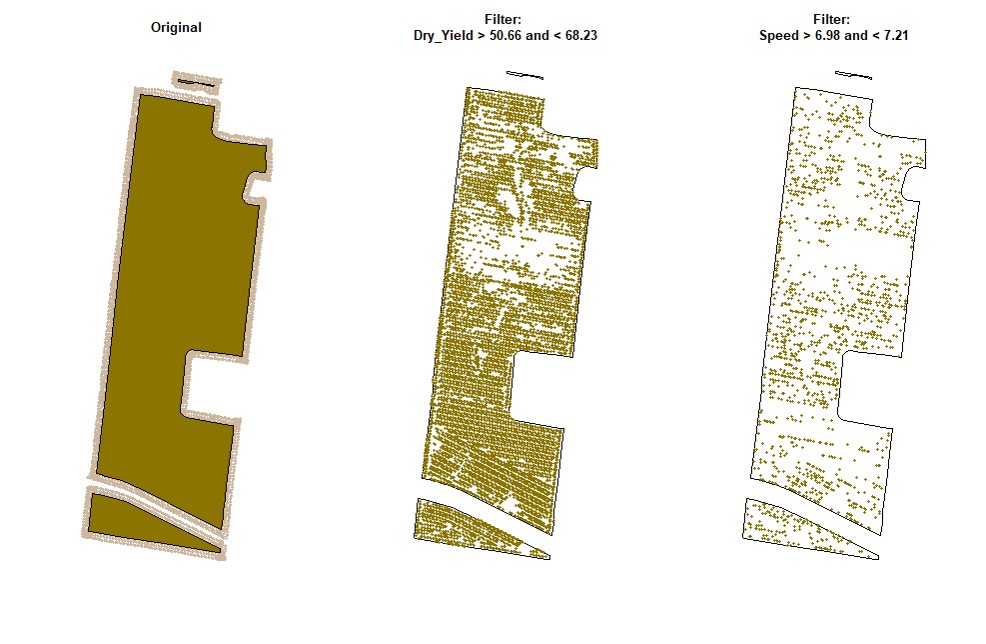
cropAbove = c(T,F))Filtering data can also be performed using standard deviation values for different traits. Function
sdField.
# Filtering data for Dry_Yield sd<0.2:
EX1.SD<-sdField(field = EX1,
trait = c("Dry_Yield"),
value = 0.2)# Filtering data for Dry_Yield sd<0.5 and Dry_Yield sd<0.2:
EX1.SD<-sdField(field = EX1,
trait = c("Dry_Yield","Dry_Yield"),
value = c(0.5,0.2))# Filtering data for Dry_Yield sd<0.5 and Speed sd<0.2 (only for the data on the small shapefile):
EX1.SD<-sdField(field = EX1,
shape = EX1.C$shape,
trait = c("Dry_Yield","Speed"),
value = c(0.5,0.2))# Filtering data for Dry_Yield sd<0.5 and Speed sd<0.2 (using the buffer shapefile):
EX1.SD<-sdField(field = EX1,
shape = EX1.B$newShape,
trait = c("Dry_Yield","Speed"),
value = c(0.5,0.2))Download and unzip the projected data example below here Parallel_Example.zip.
################
### Parallel ###
################
# General packages
library(cleanRfield)
library(terra)
# Required packages
library(parallel)
library(foreach)
library(doParallel)
# Files names (folder directory: "./field/" and "./boundary/")
field<-unique(do.call(rbind,strsplit(list.files("./field/"),split = "[.]"))[,1])
boundary<-unique(do.call(rbind,strsplit(list.files("./boundary/"),split = "[.]"))[,1])
# General filter information:
buffer=-50 # Boundary buffer of 50 feet
trait = c("Dry_Yield","Speed") # Filtered traits
filter.value = c(50,7) # cropping filter values
cropAbove = c(T,T) # All values above the filter.value
sd.value = c(1,1) # All values between sd=1
# Number of cores
n.core<-3
# Starting parallel
cl <- makeCluster(n.core, output = "")
registerDoParallel(cl)
Filtered_Field <-foreach(i = 1:length(field),
.packages = c("terra","cleanRfield")) %dopar% {
# Uploading data and boundary
F.ex<-vect(paste("./field/",field[i],".shp",sep=""))
B.ex<-vect(paste("./boundary/",boundary[i],".shp",sep=""))
# Filtering the borders by buffering the boundary shape file:
B.ex<-bufferField(shape = B.ex,value = buffer)
# Filtering data based on observed traits values:
F.ex<-filterField(field = F.ex,
shape = B.ex,
trait = trait,
value = filter.value,
cropAbove = cropAbove)
# Filtering data based on standard deviation values:
F.ex<-sdField(field = F.ex,
shape = B.ex,
trait = trait,
value = sd.value)
# New filtered data and boundary files:
return(list(NewField=wrap(F.ex), NewBoundary=wrap(B.ex)))
}
stopCluster(cl)
names(Filtered_Field)<-field
# Output
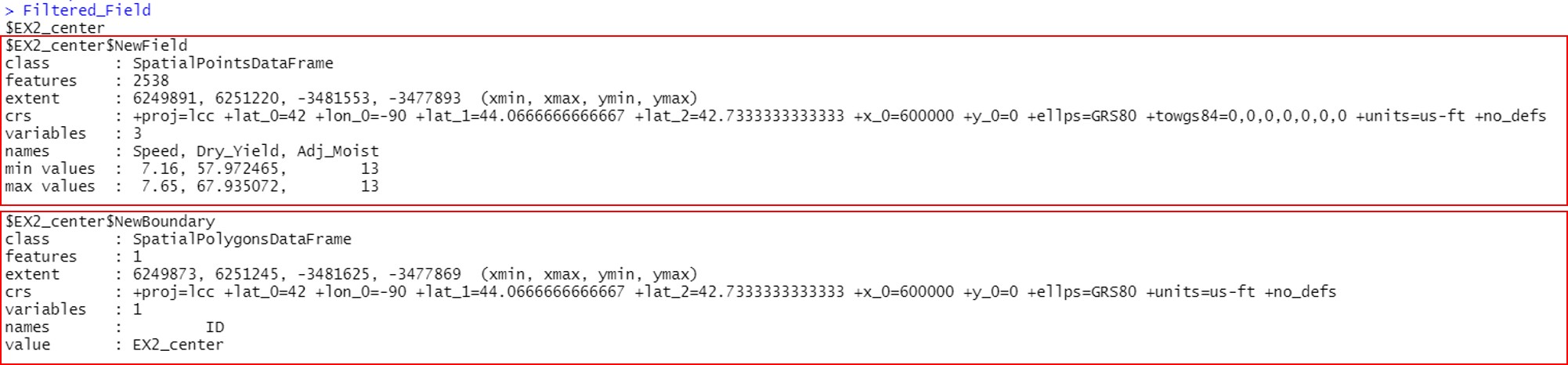
Filtered_Field <-lapply(unlist(Filtered_Field), unwrap)
Filtered_Field
# New filtered - EX2_center
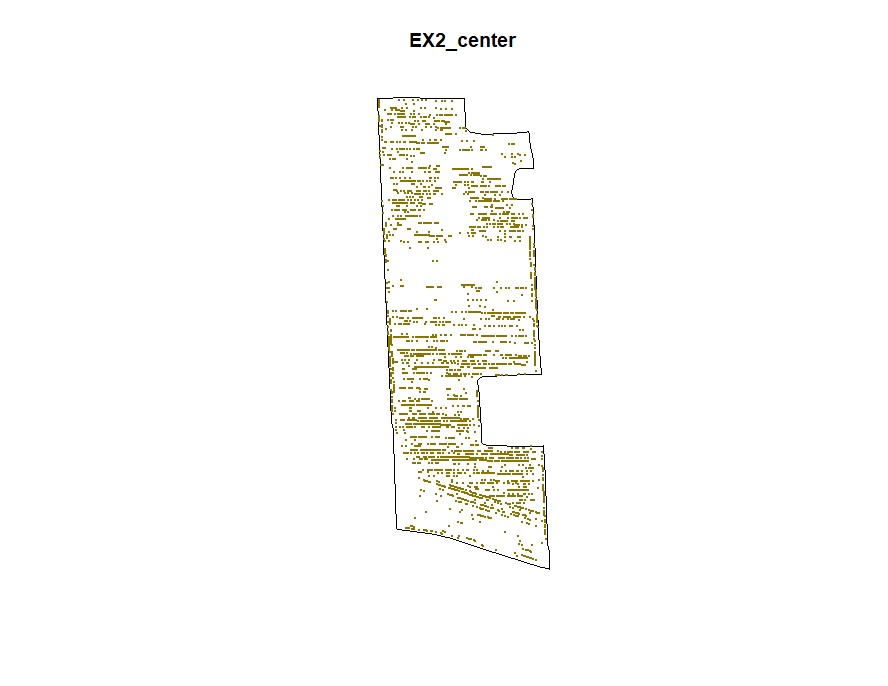
plot(Filtered_Field$EX2_center.NewBoundary, main="EX2_center")
plot(Filtered_Field$EX2_center.NewField, add=T, col="gold4",pch=20,cex=0.5)
# New filtered - EX2_north
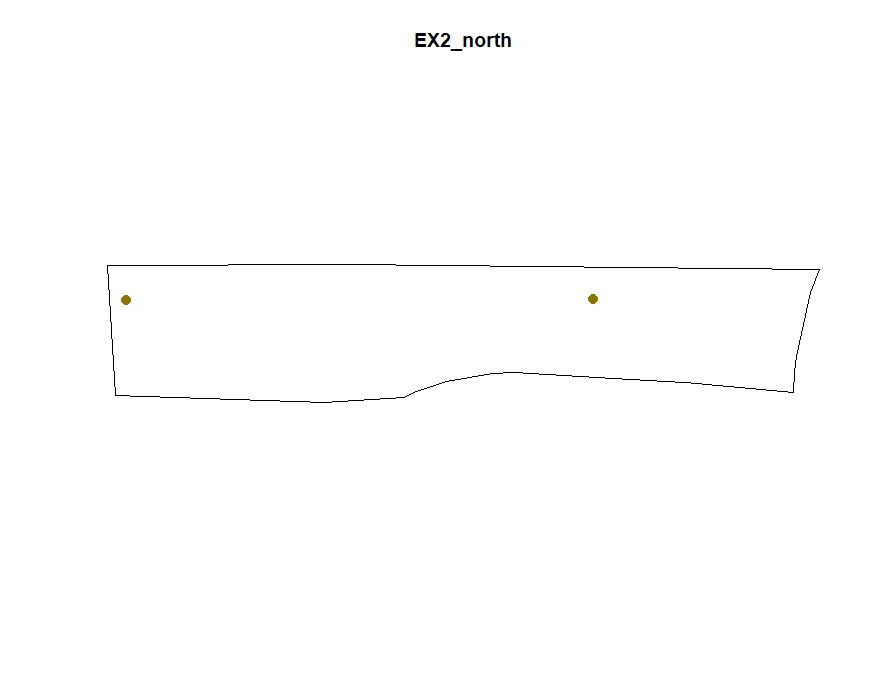
plot(Filtered_Field$EX2_north.NewBoundary, main="EX2_north")
plot(Filtered_Field$EX2_north.NewField, add=T, col="gold4",pch=20,cex=2)
# New filtered - EX2_south
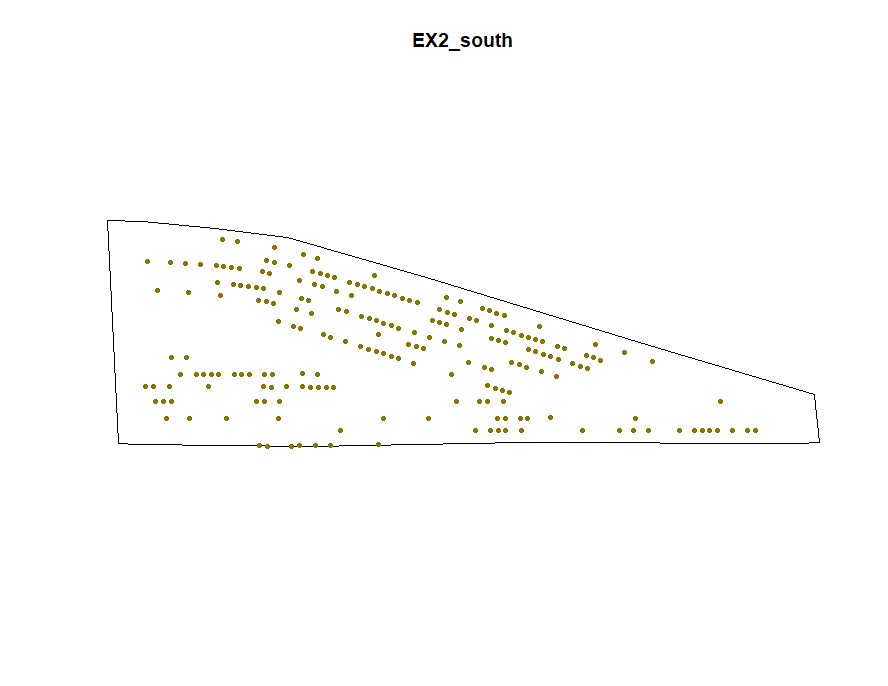
plot(Filtered_Field$EX2_south.NewBoundary, main="EX2_south")
plot(Filtered_Field$EX2_south.NewField, add=T, col="gold4",pch=20,cex=1)
# Combined new data:
NewField<-rbind(Filtered_Field$EX2_center.NewField,
Filtered_Field$EX2_north.NewField,
Filtered_Field$EX2_south.NewField)
# Giving names to each field:
Filtered_Field$EX2_center.NewBoundary$ID<-field[1]
Filtered_Field$EX2_north.NewBoundary$ID<-field[2]
Filtered_Field$EX2_south.NewBoundary$ID<-field[3]
# Combining field on the same shape file:
NewBoundary<-rbind(Filtered_Field$EX2_center.NewBoundary,
Filtered_Field$EX2_north.NewBoundary,
Filtered_Field$EX2_south.NewBoundary)
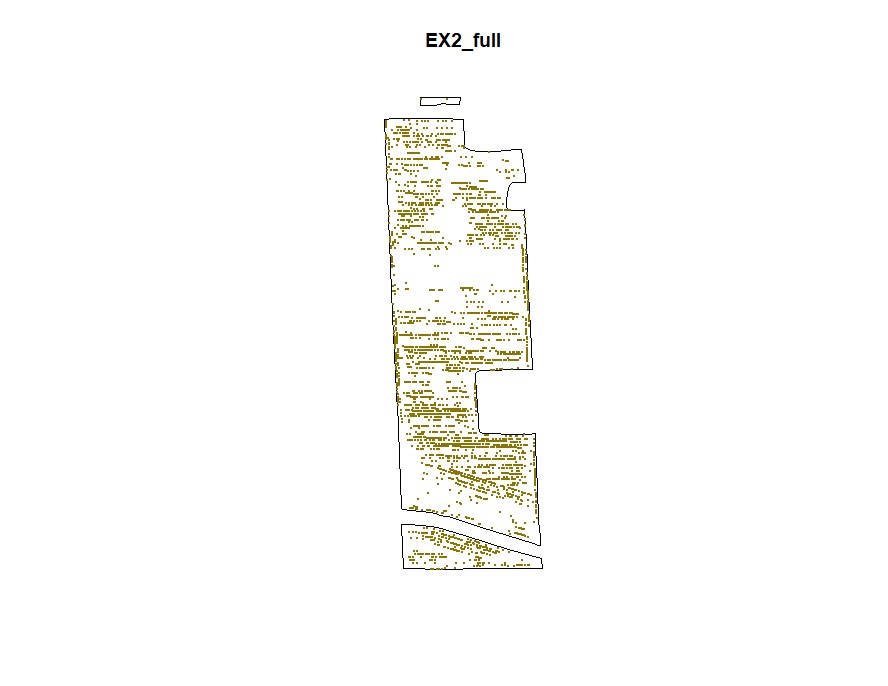
plot(NewBoundary, main="EX2_full")
plot(NewField, add=T, col="gold4",pch=20,cex=0.5)
- This example code uses the function
spplot()from the sp package to visualze "SpatialPointsDataFrames" in the plot viewing pane in R studio. The demonstrated code is useful for visualizing data before or after filtering using cleanRfield.
# Make a very basic plot where brighter colors denote higher yield
terra::plot(EX1, "Dry_Yield")
#Adjusting cuts changes the number of categories in the legend
terra::plot(EX1, "Dry_Yield", breaks=6) - If you prefer making visualizations using the package ggplot2 , we recommend converting the data from "SpatialPointsDataFrames" to "sf" objects.
#convert the object EX1 into an sf object named EX1sf
library(sf)
EX1sf<-st_as_sf(EX1)
#plot the data using geom_sf and the ggplot2 default color gradient
library(ggplot2)
ggplot()+
geom_sf(data=EX1sf, aes(color= Dry_Yield))+
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
#or make a figure using fewer of the ggplot2 display defaults
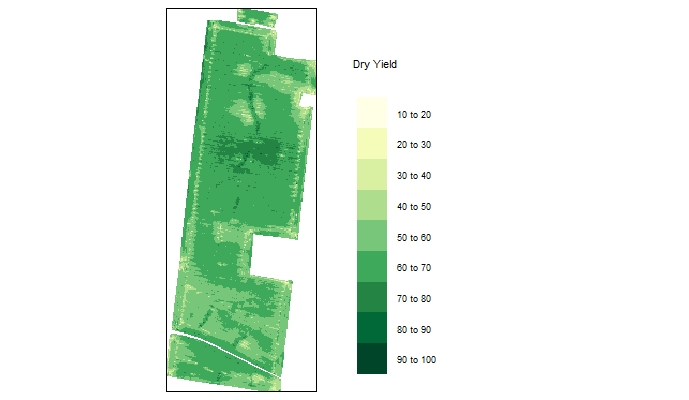
EX1.F10<-filterField(field = EX1, #filtering with Dry_Yield>10 to create a different example for plotting data
trait = "Dry_Yield",
value = 10,
cropAbove = T)
EX1sf10<-st_as_sf(EX1.F10) #converting the object EX1.F10 into an sf object
ggplot() +
geom_sf(data = EX1sf10, aes(color = Dry_Yield), size = 0.01) + #made the individual points smaller
scale_color_gradient(low = "yellow2", high = "green4") + #created a different color gradient
ggtitle("Field EX1.F10 Filtered Yield") + #added a main figure title
labs(color='Dry Yield (bu/acre)') + #changed legend title
theme_void() #removed grid background from figure
- This example code uses the function
writeVector()from the terra package to save "SpatVecor".
library(terra)
# New boundary (buffered) (SpatVector):
writeVector(EX1.B, "EX1.Buffered.shp", filetype="ESRI Shapefile")
EX1.newField <- vect("EX1.Buffered.shp") # Reading the saved boundary file.
# New point data (sampled):
writeVector(EX1.S, "EX1.5percent.shp", filetype = "ESRI Shapefile")
EX1.S.5pct <- vect("EX1.5percent.shp") # Reading the saved point file.If your data is stored as .csv or other file types, you can still utilizer cleanRfield by reading the data into a data frame in R before converting the data frame to a Spatial Points Data frame. This example uses a .csv file as the data source, but any data frame object in R that has coordinates can be converted to a spatial points data frame using this method regardless of data source file type. This data is in latitude and longitude (unprojected data). You will need to use a different CRS in the proj4string section if your data is projected. See the example code below and learn more about SpatialPoints in the documentation for the package sp.
Download the example: EX3.csv
library(terra)
# reading in the .csv file
DF<-read.csv("EX3.csv")
colnames(DF) #checking that the latitude is the first column and longitude is the second column
# creating the coordinates object using the latitude and longigude columns
xy <- DF[,c(1,2)]
# creating a new spatial points data frame from the data in DF
SpatialDF <- vect(DF, geom=c('Long','Lat'), crs = "+proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0")
crs(SpatialDF)Users may want to interpolate their yield observations to create a raster data set for visualization or further data analysis. Below, we have provided some example code for interpolating yield maps using either inverse distance weighting (IDW) or ordinary kriging. In general, we recommend IDW due to its faster processing time for large data sets.
In this example code, we use the same example data (EX1) as in tutorial section 1. You may also need to install the package tmap before proceeding with the provided code.
The next code section provides an example for running IDW in R. Users will load the required pacakges, load and filter yield data, transform filtered data and the field boundary file, prepare an empty grid, run the IDW interpolation, and finally make a map to visualize the interpolation. Transformation is a step included in this workflow since the EX1 shapefile is not in a projected CRS, and transforming into a projected CRS helps align the yield map observations to the empty grid. Preparing the empty grid is necessary to determine the extent and resolution of the interpolation.
#### packages to run basic filtering with cleanRfield ####
library(terra)
library(cleanRfield)
#### additional packages needed for interpolation & mapping ####
library(gstat) # used to make the idw model
library(sf) # used to convert spatial objects
library(sp) # used to prepare the raster grid with spsample function
library(tmap) # used for visualization
#### preparing the yield data ####
EX1 <- vect("EX1.shp") # EX1.shp download link is in tutorial section 1
EX1.Shape <- vect("EX1_boundary.shp") #EX1_boundary.shp download link is in tutorial section 1
# filtering data to remove biologically unlikely soybean yield observations and NA values
EX1.F <- filterField(field = EX1,
trait = c("Dry_Yield","Dry_Yield"),
value = c(10,100),
cropAbove = c(T,F))
# transforming the filtered data so that it is a projected CRS
EX1_merc <- spTransform(as_Spatial(st_as_sf(EX1.F)), CRS=CRS("+proj=merc +ellps=GRS80"))
EX1_merc # looking at summary output to check projection
#transforming the boundary too-- this will be used later for visualization
EX1.Shape_merc <- spTransform(as_Spatial(st_as_sf(EX1.Shape)), CRS=CRS("+proj=merc +ellps=GRS80"))
EX1.Shape_merc #looking at summary output to check projection
#### preparing an empty grid ####
G <- as.data.frame(spsample(EX1_merc, "regular", n=50000)) #n = total number of grid cells
names(G) <- c("X", "Y")
coordinates(G) <- c("X", "Y")
gridded(G) <- TRUE # create SpatialPixel object
fullgrid(G) <- TRUE # create SpatialGrid object
proj4string(G) <- proj4string(EX1_merc) # using the projection from EX1_merc to project the grid G
proj4string(G) # checking that G is projected
#### running IDW using the yield data and empty grid ####
Yield.idw <- gstat::idw(Dry_Yield ~ 1, EX1_merc, newdata=G, idp=2.0)
#### visualizing IDW interpolation ####
r.idw <- raster::raster(Yield.idw) # convert the IDW model to a RasterStack
r.masked <- raster::mask(r.idw, EX1.Shape_merc) # mask the raster to the field boundary
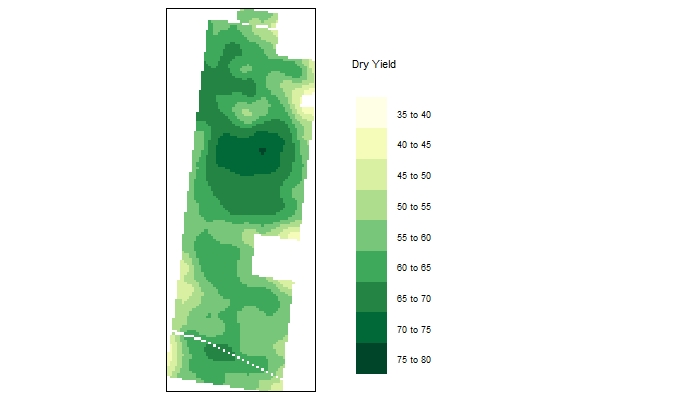
yieldmap.idw <- tm_shape(r.masked) + #make the map using functions from the tmap library
tm_raster(n=10,palette = "YlGn",
title="Dry Yield") +
tm_legend(legend.outside=TRUE)
yieldmap.idw #view the map
The following codes sections provides an example for interpolating via ordinary kriging in R. This workflow begins very similarly to the IDW interpolation workflow until we begin creating variogram models
#### packages to run basic filtering with cleanRfield ####
library(terra)
library(cleanRfield)
#### additional packages needed for interpolation & mapping ####
library(gstat) # used to make the idw model
library(sf) # used to convert spatial objects
library(sp) # used to prepare the raster grid with spsample function
library(tmap) # used for visualization
#### preparing the yield data ####
EX1 <- vect("EX1.shp") # EX1.shp download link is in tutorial section 1
EX1.Shape <- vect("EX1_boundary.shp") #EX1_boundary.shp download link is in tutorial section 1
# filtering data to remove biologically unlikely soybean yield observations and NA values
EX1.F <- filterField(field = EX1,
trait = c("Dry_Yield","Dry_Yield"),
value = c(10,100),
cropAbove = c(T,F))
# transforming the filtered data so that it is a projected CRS
EX1_merc <- spTransform(as_Spatial(st_as_sf(EX1.F)), CRS=CRS("+proj=merc +ellps=GRS80"))
EX1_merc # looking at summary output to check projection
# transforming the boundary too-- this will be used later for visualization
EX1.Shape_merc <- spTransform(as_Spatial(st_as_sf(EX1.Shape)), CRS=CRS("+proj=merc +ellps=GRS80"))
EX1.Shape_merc #looking at summary output to check projection
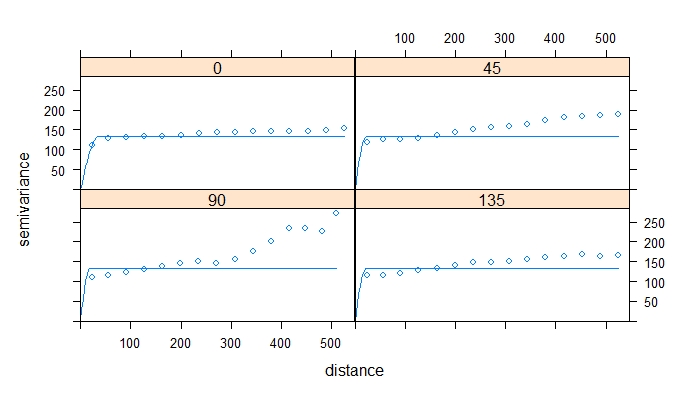
#### make a variogram to assess spatial relationships between yield observations ####
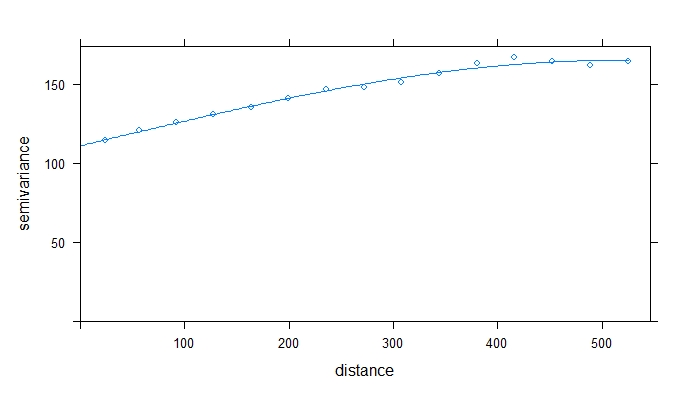
v_overall <- variogram(Dry_Yield~1, data = EX1_merc)
plot(v_overall) # visually estimate sill, model shape, range, and nugget
vmodel_overall <- vgm(psill=150, model="Sph", nugget=100, range=400) # estimate variogram model
fittedmodel_overall <- fit.variogram(v_overall, model=vmodel_overall) # fit variogram model
fittedmodel_overall #print the fitted model to see how it compares to your initial estimate
plot(v_overall, model=fittedmodel_overall)
The example variogram above had a range of ~509m, which indicates that yield observations that are <509m apart are spatially correlated. If you want to learn more about variograms, try this blog post from GIS Geography. Next, we will check that the data is not anisotrophic by developing 4 separate directional variograms.
# let's see if the spatial autocorrelation is the same in all directions-- is the data anisotrophic?
gs_object <- gstat(formula=Dry_Yield~ 1, data=EX1_merc)
v_directional <- variogram(gs_object, alpha=c(0,45,90,135))
vmodel_directional <- vgm(model='Sph' , anis=c(0, 0.5))
fittedmodel_directional <- fit.variogram(v_directional, model=vmodel_directional)
plot(v_directional, model=fittedmodel_directional, as.table=TRUE)
There is not a commonly applied statistical test for anisotrophy, so this decision is a judgement call that each person will make a little differently. For distances <250m, these models are pretty similar. the data is not perfectly stationary, but in our judgement, it is not so anisotrophic that kriging would be inappropriate. If you perform directional variograms and there are very substantial differences between the models, we do not recommend kriging for interpolation. Instead, try another interpolation method that doesn't assume stationarity
Kriging takes a long time to compute, so in this example we will randomly sample 20% the yield observations before kriging to save time. Depending on your computer and your use for the kriged map, you may want to sample even fewer points, or krige using all observations. In this example we also used a lower resolution empty grid than in the IDW example to save computational time.
#### sample 20% of yield observations ####
EX1_merc_10pct<-sampleField(field = EX1_merc, size = 0.2)
#### update the empty grid and gstat object ####
# prepare a similar, but smaller, empty grid than the IDW example code
G <- as.data.frame(spsample(EX1_merc, "regular", n=10000))
names(G) <- c("X", "Y")
coordinates(G) <- c("X", "Y")
gridded(G) <- TRUE # Create SpatialPixel object
fullgrid(G) <- TRUE # Create SpatialGrid object
proj4string(G) <- proj4string(EX1_merc)
proj4string(G) #checking that G is projected
# now update the gstat object from before so that it includes the fitted
# model, not the estimated model from earlier in the kriging workflow
gs_object <- gstat(formula=Dry_Yield~ 1,
data=EX1_merc_10pct, model=fittedmodel_overall)
#### run the kriging procedure using the gstat object and empty grid ####
kriged_surface <- predict(gs_object, model=fittedmodel_overall, newdata=G)
summary(kriged_surface)
There's a reason most point-and-click softwares highly recommend not kriging yield maps. Even with just 20% of the observations, that took almost 5 min to run on my laptop. Running 50% of the observations takes me over 25 min. Fortunately, once the kriged surface is made, visualization is quick.
#### visualizing kriged map ####
kriged_raster <- raster::raster(kriged_surface)
kriged_masked <- raster::mask(kriged_raster, EX1.Shape_merc)
tm_shape(kriged_masked) +
tm_raster(n=10,palette = "YlGn",
title="Dry Yield") +
tm_legend(legend.outside=TRUE)Regardless of interpolation method chosen, we highly recommend assessing the fit of your model using cross-validation and other methods. We do not provide code for assessing goodness of fit in this tutorial, but you can provide more information on this process from a variety of GIS tutorials. We find Manuel Gimond's tutorial to be particularly helpful.
This discussion group provides an online source of information about the cleanRfield package. Report a bug and ask a question at:
The R/cleanRfield package as a whole is distributed under GPL-2 (GNU General Public License version 2).
coming soon...