Final project presented to the Artificial Intelligence I course, with code INF 420, taught by Professor Julio C. S. Reis, as a partial requirement for approval in the course.
The proposed project is a chatbot responsible for screening in hospitals to differentiate COVID cases from other similar symptomatic cases, such as allergy, cold, and flu. Additionally, at the end, it generates a medical record of the patient with personal information, such as name, age, occupation, symptoms, the number of days the person has been sick, and the result of the prediction made using artificial intelligence techniques.
To use the chatbot, it is necessary to run randomforest_knn.py and trainingBot.py before running chatBot.py.
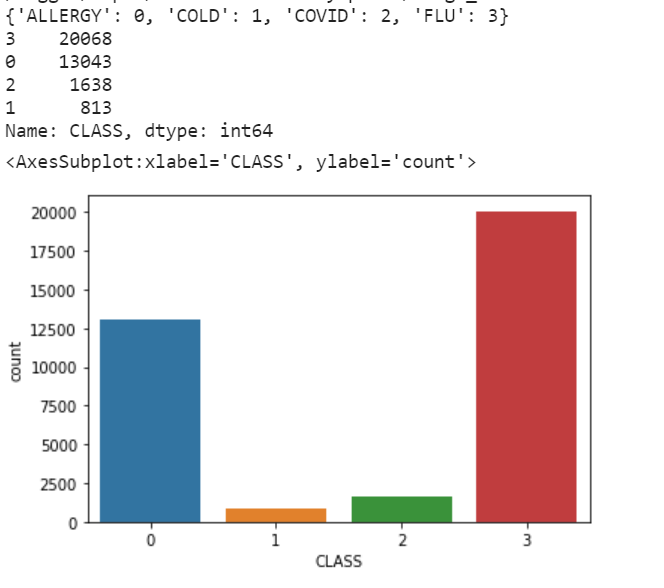
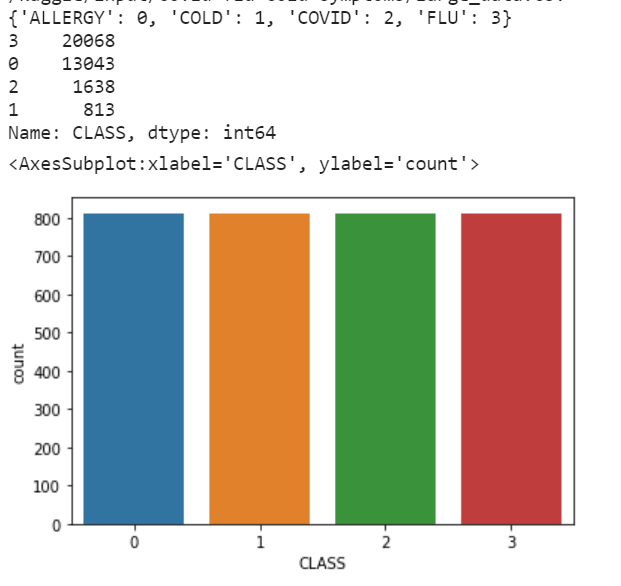
I used a dataset available on Kaggle, containing various illness cases along with their symptoms and the classification of each case as allergy, cold, COVID, or flu. This database is imbalanced, with many cases of allergy and flu compared to COVID and cold. This is a problem as it can lead to many "false alarms," with the diagnosis leaning heavily towards allergy and flu due to their higher sample count. The symptoms among the four illnesses are quite similar, making it challenging to assess effectiveness. Therefore, I employed the under-sampling technique using the RandomUnderSampler() method from the imblearn.under_sampling library.
To address the classification problem, I employed two techniques: Random Forest and K-Nearest Neighbor. For Random Forest, I set the number of decision trees to 10, with separation criterion as entropy, and a random_state of 42. For K-Nearest Neighbor, I chose a number of neighbors as 10 and used the Euclidean metric. The training of the models is in the file randomforest_knn.py. After training, the program automatically saves the models, with the assistance of the joblib library, as they will be used for classification in the chatbot.py file.
For the development of the chatbot, I created a simple neural network with three dense layers and two dropout layers. I used Adam as the optimizer with a learning rate of 0.01, as it was the best optimizer I had found when working on assignment 4 of this course (I also tested with SGD, but Adam showed better performance). The training of the chatbot is done in the trainingBot.py file, and the application of the chatbot is done in the chatBot.py file. I had to implement the chatbot in English because I used the WordLemmatizer method from the nltk library, which groups inflected forms of a word so that they can be analyzed as a single item. For example, in English, we have various variations of the word 'work', such as 'works,' 'worked,' 'working.' This method combines all these forms to be analyzed as 'work.' I didn't find an easy way to use this method in Portuguese, so I apologize if you find any errors in English.
To conclude, the chatbot.py program, after collecting all the data, such as personal information and symptoms of the patient, generates a docx file, which serves as the patient's medical record. .
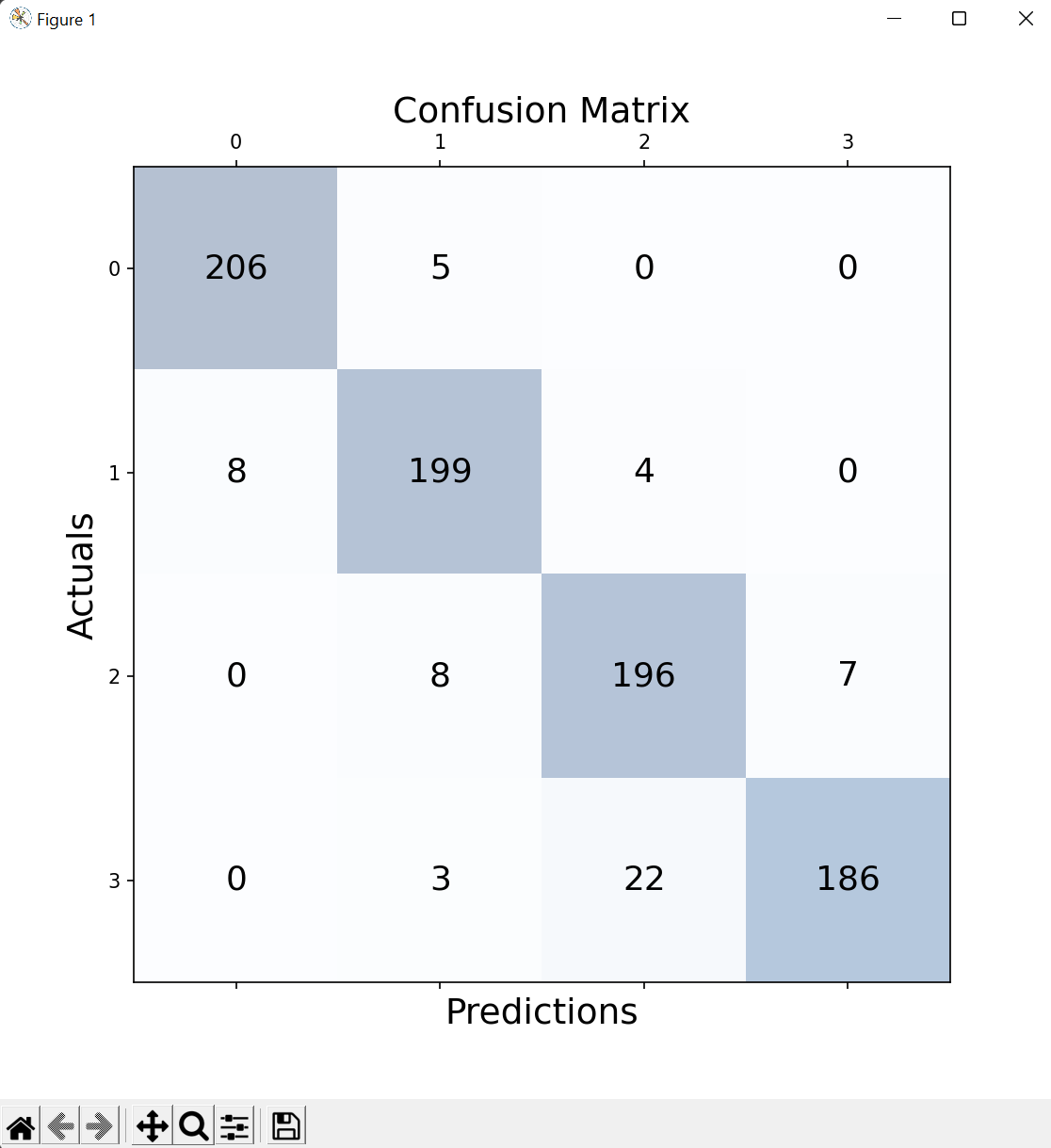
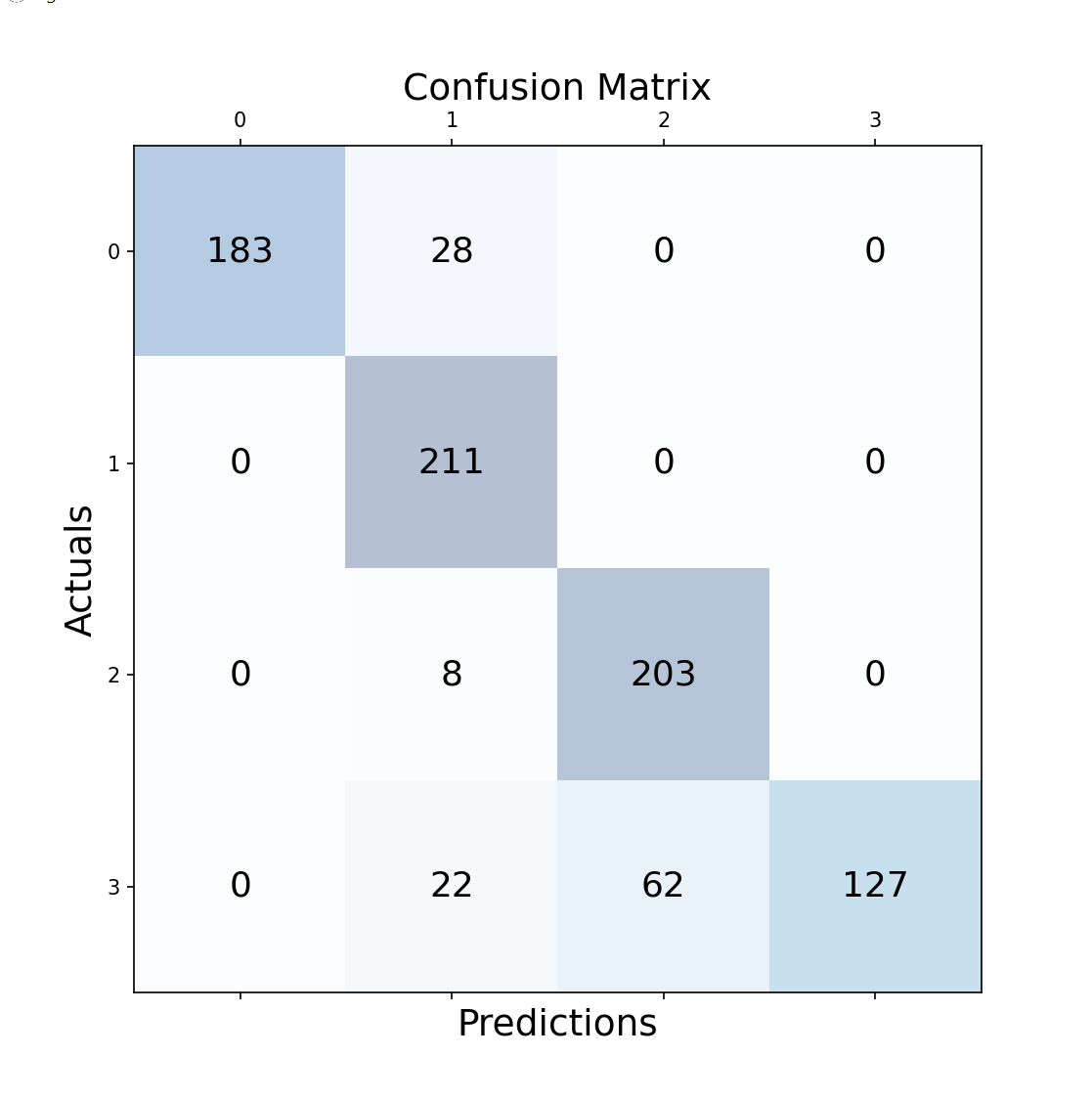
Regarding KNN and Random Forest, I chose multiple metrics to analyze effectiveness, ensuring we don't rely on just one and potentially have a false impression of the model's performance. Remembering that: {0: Allergy, 1: Cold, 2: COVID, 3: Flu}
Random Forest:With an average model accuracy of 93%, we can observe in the confusion matrix that true positives and true negatives occur much more frequently than false positives and false negatives. Cases of flu, which have a higher tendency to be easily confused with COVID, are more prone to errors.
---------------- Random Forest ----------------
precision recall f1-score support
0 0.96 0.98 0.97 211
1 0.93 0.94 0.93 211
2 0.88 0.93 0.91 211
3 0.96 0.88 0.92 211
accuracy 0.93 844
macro avg 0.93 0.93 0.93 844
weighted avg 0.93 0.93 0.93 844
K-Nearest neighbor:TWith an average model accuracy of 86%, we can observe in the confusion matrix that true positives and true negatives occur much more frequently than false positives and false negatives. Cases of flu, which have a higher tendency to be easily confused with COVID and colds, are more prone to errors.
---------------- K-Nearest neighbor ----------------
precision recall f1-score support
0 1.00 0.87 0.93 211
1 0.78 1.00 0.88 211
2 0.77 0.96 0.85 211
3 1.00 0.60 0.75 211
accuracy 0.86 844
macro avg 0.89 0.86 0.85 844
weighted avg 0.89 0.86 0.85 844
For the neural network used in building the chatbot, we achieved an accuracy of 98.25% after training for 200 epochs.
Epoch 1/100
12/12 [==============================] - 1s 2ms/step - loss: 2.0453 - accuracy: 0.2281
Epoch 2/100
12/12 [==============================] - 0s 2ms/step - loss: 1.7778 - accuracy: 0.4035
Epoch 3/100
12/12 [==============================] - 0s 2ms/step - loss: 1.4486 - accuracy: 0.5088
Epoch 4/100
12/12 [==============================] - 0s 2ms/step - loss: 1.0473 - accuracy: 0.6842
Epoch 5/100
12/12 [==============================] - 0s 2ms/step - loss: 0.7694 - accuracy: 0.7544
Epoch 6/100
12/12 [==============================] - 0s 3ms/step - loss: 0.6967 - accuracy: 0.7368
Epoch 7/100
12/12 [==============================] - 0s 2ms/step - loss: 0.4201 - accuracy: 0.8596
Epoch 8/100
12/12 [==============================] - 0s 4ms/step - loss: 0.2586 - accuracy: 0.9474
Epoch 9/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2749 - accuracy: 0.9298
Epoch 10/100
12/12 [==============================] - 0s 2ms/step - loss: 0.3427 - accuracy: 0.8772
Epoch 11/100
12/12 [==============================] - 0s 1ms/step - loss: 0.2164 - accuracy: 0.9123
Epoch 12/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1798 - accuracy: 0.9298
Epoch 13/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2381 - accuracy: 0.9123
Epoch 14/100
12/12 [==============================] - 0s 1ms/step - loss: 0.2199 - accuracy: 0.8596
Epoch 15/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2347 - accuracy: 0.9474
Epoch 16/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1487 - accuracy: 0.9825
Epoch 17/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1959 - accuracy: 0.9474
Epoch 18/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1510 - accuracy: 0.9474
Epoch 19/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1690 - accuracy: 0.9474
Epoch 20/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1701 - accuracy: 0.9298
Epoch 21/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0837 - accuracy: 0.9649
Epoch 22/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0232 - accuracy: 1.0000
Epoch 23/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0664 - accuracy: 0.9649
Epoch 24/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0539 - accuracy: 0.9649
Epoch 25/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0805 - accuracy: 0.9649
Epoch 26/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1295 - accuracy: 0.9649
Epoch 27/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1153 - accuracy: 0.9649
Epoch 28/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2349 - accuracy: 0.9474
Epoch 29/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1176 - accuracy: 0.9825
Epoch 30/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0626 - accuracy: 0.9825
Epoch 31/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2018 - accuracy: 0.9474
Epoch 32/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2196 - accuracy: 0.9649
Epoch 33/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2463 - accuracy: 0.9649
Epoch 34/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1690 - accuracy: 0.9474
Epoch 35/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2359 - accuracy: 0.9474
Epoch 36/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1023 - accuracy: 0.9825
Epoch 37/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0949 - accuracy: 0.9825
Epoch 38/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1134 - accuracy: 0.9649
Epoch 39/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1805 - accuracy: 0.9298
Epoch 40/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0570 - accuracy: 0.9825
Epoch 41/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0529 - accuracy: 0.9825
Epoch 42/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0832 - accuracy: 0.9649
Epoch 43/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0331 - accuracy: 1.0000
Epoch 44/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0498 - accuracy: 0.9649
Epoch 45/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0491 - accuracy: 0.9825
Epoch 46/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0723 - accuracy: 0.9474
Epoch 47/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0846 - accuracy: 0.9474
Epoch 48/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0933 - accuracy: 0.9474
Epoch 49/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0469 - accuracy: 0.9825
Epoch 50/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0543 - accuracy: 0.9825
Epoch 51/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0387 - accuracy: 0.9825
Epoch 52/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1329 - accuracy: 0.9474
Epoch 53/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0476 - accuracy: 1.0000
Epoch 54/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0939 - accuracy: 0.9474
Epoch 55/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0512 - accuracy: 0.9825
Epoch 56/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1091 - accuracy: 0.9474
Epoch 57/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0597 - accuracy: 0.9825
Epoch 58/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0659 - accuracy: 0.9649
Epoch 59/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0336 - accuracy: 0.9825
Epoch 60/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0988 - accuracy: 0.9474
Epoch 61/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0961 - accuracy: 0.9649
Epoch 62/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0660 - accuracy: 0.9474
Epoch 63/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2421 - accuracy: 0.9123
Epoch 64/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0685 - accuracy: 0.9649
Epoch 65/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1687 - accuracy: 0.9474
Epoch 66/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0682 - accuracy: 0.9474
Epoch 67/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0365 - accuracy: 0.9825
Epoch 68/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0474 - accuracy: 1.0000
Epoch 69/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0780 - accuracy: 0.9649
Epoch 70/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0607 - accuracy: 0.9649
Epoch 71/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0730 - accuracy: 0.9649
Epoch 72/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0543 - accuracy: 0.9649
Epoch 73/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0328 - accuracy: 0.9825
Epoch 74/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1422 - accuracy: 0.9649
Epoch 75/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0918 - accuracy: 0.9649
Epoch 76/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1991 - accuracy: 0.9474
Epoch 77/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0222 - accuracy: 0.9825
Epoch 78/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1473 - accuracy: 0.9298
Epoch 79/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1072 - accuracy: 0.9649
Epoch 80/100
12/12 [==============================] - 0s 1ms/step - loss: 0.0910 - accuracy: 0.9825
Epoch 81/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0554 - accuracy: 0.9649
Epoch 82/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1140 - accuracy: 0.9649
Epoch 83/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0929 - accuracy: 0.9825
Epoch 84/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0358 - accuracy: 0.9825
Epoch 85/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2528 - accuracy: 0.9123
Epoch 86/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0476 - accuracy: 0.9825
Epoch 87/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0566 - accuracy: 0.9649
Epoch 88/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0430 - accuracy: 0.9649
Epoch 89/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0245 - accuracy: 1.0000
Epoch 90/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0595 - accuracy: 0.9649
Epoch 91/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0246 - accuracy: 0.9825
Epoch 92/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0368 - accuracy: 0.9825
Epoch 93/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0187 - accuracy: 1.0000
Epoch 94/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1179 - accuracy: 0.9474
Epoch 95/100
12/12 [==============================] - 0s 2ms/step - loss: 0.2349 - accuracy: 0.9298
Epoch 96/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0602 - accuracy: 0.9649
Epoch 97/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0887 - accuracy: 0.9649
Epoch 98/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1035 - accuracy: 0.9649
Epoch 99/100
12/12 [==============================] - 0s 2ms/step - loss: 0.0724 - accuracy: 0.9474
Epoch 100
/100
12/12 [==============================] - 0s 2ms/step - loss: 0.1765 - accuracy: 0.9825
Training complete
Despite the models having high accuracies, there are many cases of flu and colds that are confused with COVID. Since COVID is a highly contagious disease that caused a pandemic affecting the entire world, it might not be advisable to rely too heavily on the diagnosis of COVID, flu, and colds, as they are confused more frequently than I expected. However, the models were excellent at distinguishing these cases from allergies, which is beneficial because allergies have symptoms very similar to COVID.
https://sigmoidal.ai/como-lidar-com-dados-desbalanceados/
https://www.youtube.com/watch?v=1lwddP0KUEg&t=1737s