Score the regional preference for scRNAseq cluster based on regional bulkRNAseq data.
You can install the development version of ScoreRegionPreference from GitHub with:
# install.packages("devtools")
devtools::install_github("fentouxungui/ScoreRegionPreference")This is a basic example which shows you how to solve a common problem:
library(Seurat)
#> Warning: 程辑包'Seurat'是用R版本4.3.1 来建造的
#> The legacy packages maptools, rgdal, and rgeos, underpinning the sp package,
#> which was just loaded, will retire in October 2023.
#> Please refer to R-spatial evolution reports for details, especially
#> https://r-spatial.org/r/2023/05/15/evolution4.html.
#> It may be desirable to make the sf package available;
#> package maintainers should consider adding sf to Suggests:.
#> The sp package is now running under evolution status 2
#> (status 2 uses the sf package in place of rgdal)
#> Attaching SeuratObject
library(gridExtra)
library(ScoreRegionPreference)使用EC的各个区段的RNA-seq值,来对单细胞中的各个EC细胞亚群,进行定位预测。
单细胞数据来自文章: Hung R J, Hu Y, Kirchner R, et al. A cell atlas of the adult Drosophila midgut[J]. Proceedings of the National Academy of Sciences, 2020, 117(3): 1514-1523.
RNA-seq数据来自Flygut-seq: Cell and region specific gene expression of the fly midgut
将RNAseq里的基因ID转为symbol,注意,要使用与单细胞数据分析用的GTF文件来生成FlyGeneMeta。
# RNAseq data
data(FlyGeneMeta)
data(RNAseq)
head(RNAseq$EC)
#> R1 R2 R3 R4 R5
#> FBgn0000003 0.00000000 0.00000000 0.00000000 0.00000000 0.00000000
#> FBgn0000008 0.99406575 1.96251049 0.31337624 1.90011232 0.52664174
#> FBgn0000014 0.02751965 0.14201820 0.10356404 0.30602071 0.15670007
#> FBgn0000015 0.01164936 0.08265733 0.05089256 0.17023985 0.09432941
#> FBgn0000017 0.01560269 0.04168749 0.03383766 0.05107372 0.04774208
#> FBgn0000018 4.01871689 4.47380060 4.42188673 4.79100658 4.35939753
bulkRNAseq <- scRNAseq_Score_Region_Check(RNAseq$EC, FlyGeneMeta)
#> 395 features from data frame not exist in meta file!
head(bulkRNAseq)
#> R1 R2 R3 R4 R5
#> 7SLRNA:CR32864 0.00000000 0.00000000 0.00000000 0.00000000 0.00000000
#> a 0.99406575 1.96251049 0.31337624 1.90011232 0.52664174
#> abd-A 0.02751965 0.14201820 0.10356404 0.30602071 0.15670007
#> Abd-B 0.01164936 0.08265733 0.05089256 0.17023985 0.09432941
#> Abl 0.01560269 0.04168749 0.03383766 0.05107372 0.04774208
#> abo 4.01871689 4.47380060 4.42188673 4.79100658 4.35939753# scRNAseq data
data(scRNA)
scRNA <- UpdateSeuratObject(scRNA)
#> Validating object structure
#> Updating object slots
#> Ensuring keys are in the proper structure
#> Ensuring feature names don't have underscores or pipes
#> Object representation is consistent with the most current Seurat version
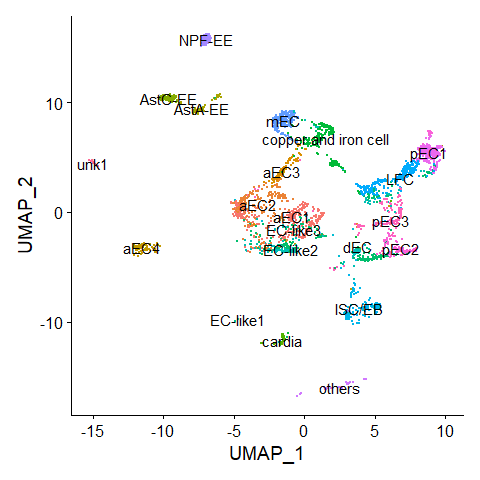
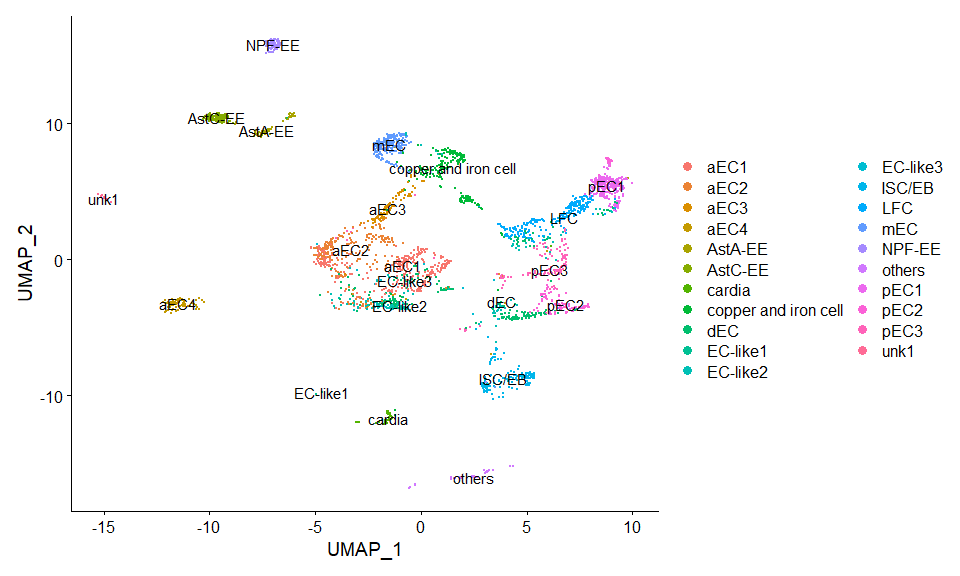
DimPlot(scRNA, label = TRUE) + NoLegend()score.list <- scRNAseq_Score_Region(scRNA, bulkRNAseq)
#> 2724 features from RNA-seq not exist in scRNAseq!
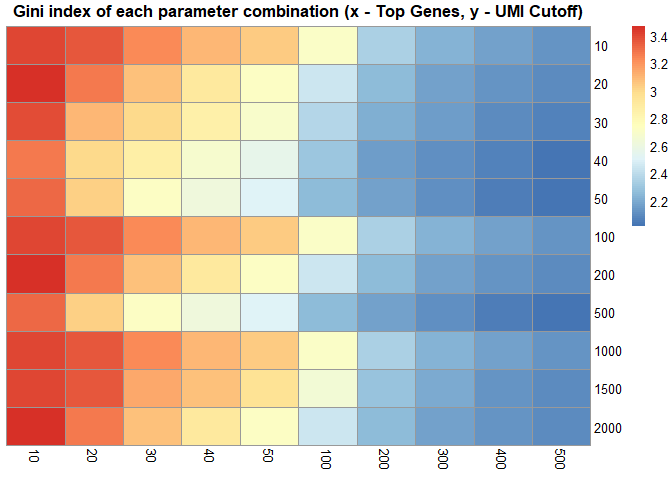
scRNAseq_Score_Region_evaluate(score.list, cluster_rows = FALSE, cluster_cols = FALSE,
main = "Gini index of each parameter combination (x - Top Genes, y - UMI Cutoff)")# correlation of each parameter combination
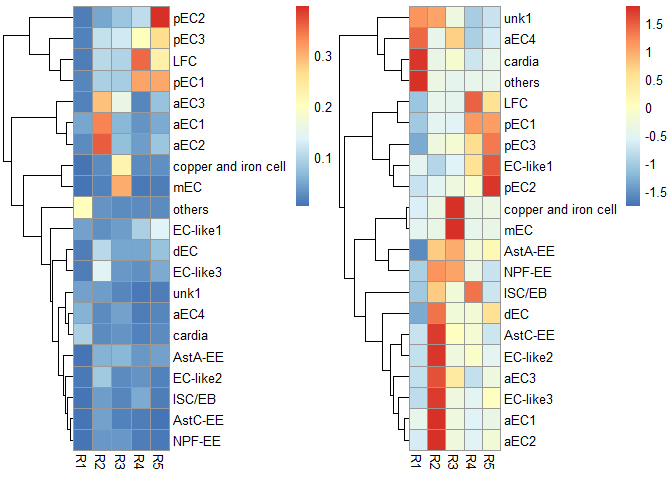
# scRNAseq_Score_Region_evaluate2(score.list)使用默认参数,即组合有最大Gini index value。
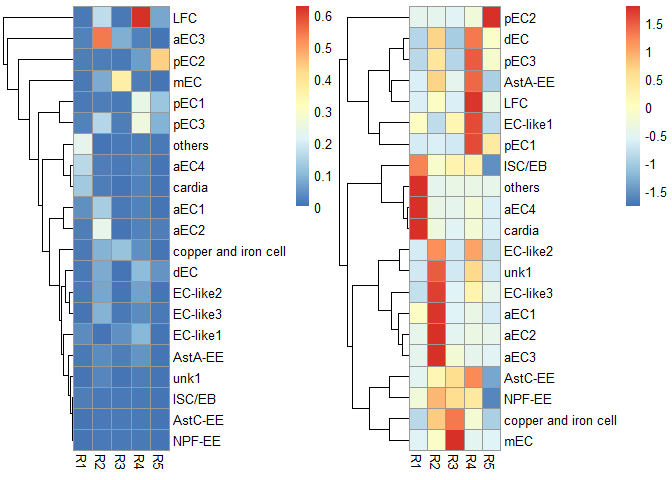
p1 <- scRNAseq_Score_Region_plot(score.list, cluster_cols = FALSE, silent = TRUE)
#> Using UMI Cutoff: 20; Genes Used: 10
p2 <- scRNAseq_Score_Region_plot(score.list, cluster_cols = FALSE, scale = "row", silent = TRUE)
#> Using UMI Cutoff: 20; Genes Used: 10
grid.arrange(p1[[4]],p2[[4]],nrow = 1) & NoLegend()#> NULL
# show the selected genes
futile.logger::flog.threshold(futile.logger::ERROR, name = "VennDiagramLogger")
#> NULL
scRNAseq_Score_Region_ExtractFeatures(score.list)
#> Using UMI Cutoff: 20; Genes Used: 10#> NULL
#> R1 R2 R3 R4 R5
#> [1,] "CG17738" "Mal-A7" "Zip42C.2" "Ser8" "CG17477"
#> [2,] "CG3819" "CG4830" "CG6277" "CG11911" "CG31265"
#> [3,] "CG6839" "Npc2f" "CG43789" "CG11912" "CG17475"
#> [4,] "CG43673" "CG8834" "Vha100-4" "CG17571" "CG4053"
#> [5,] "CG11854" "CG15617" "CG6901" "Jon99Fi" "CG6337"
#> [6,] "to" "CG9682" "CG31446" "CG10472" "CG32379"
#> [7,] "CG43134" "Peritrophin-15a" "CG31663" "Jon99Fii" "CG8774"
#> [8,] "Yp3" "CG30340" "CG5770" "CG7953" "CG31267"
#> [9,] "CG15649" "CG34040" "CG30479" "Jon66Ci" "CG43208"
#> [10,] "Muc68D" "CG18404" "CG17930" "Jon44E" "CG32284"
使用自定义参数,即设定UMI Cutoff为100和选取前100个基因。
p1 <- scRNAseq_Score_Region_plot(score.list, 100, 100, cluster_cols = FALSE, silent = TRUE)
p2 <- scRNAseq_Score_Region_plot(score.list, 100, 100, cluster_cols = FALSE, scale = "row", silent = TRUE)
grid.arrange(p1[[4]],p2[[4]],nrow = 1) & NoLegend()#> NULL
# show the selected genes
# futile.logger::flog.threshold(futile.logger::ERROR, name = "VennDiagramLogger")
# scRNAseq_Score_Region_ExtractFeatures(score.list, 100, 100)可以看到,使用这两种参数,都可以准确判定EC亚群的定位,并可以给出更准确的定位。
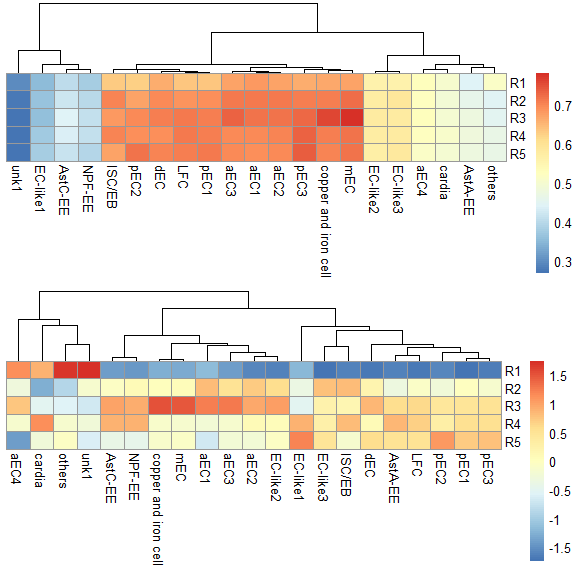
使用所有基因
score.matrix <- scRNAseq_Score_Region2(scRNA, bulkRNAseq, Method = "spearman")
#> 2724 features from RNA-seq not exist in scRNAseq!
p1 <- pheatmap::pheatmap(score.matrix, cluster_rows = FALSE, silent = TRUE)
p2 <- pheatmap::pheatmap(score.matrix, scale = "column", cluster_rows = FALSE, silent = TRUE)
grid.arrange(p1[[4]],p2[[4]],nrow = 2) & NoLegend()#> NULL
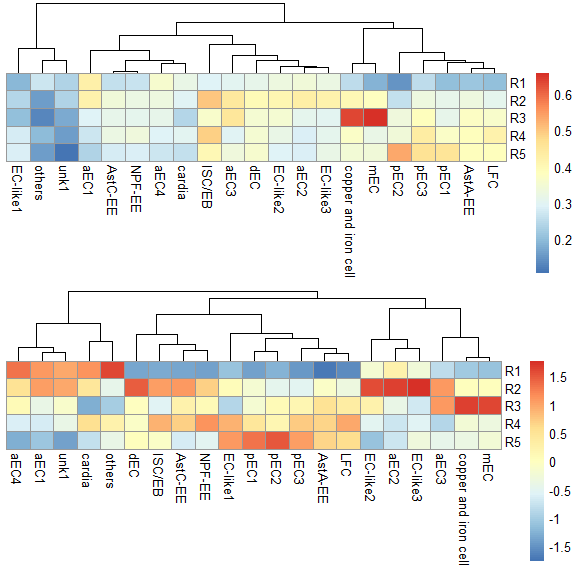
使用Top基因
score.matrix <- scRNAseq_Score_Region2(scRNA, bulkRNAseq, Method = "spearman", Genes.Selection = "Top")
#> 2724 features from RNA-seq not exist in scRNAseq!
p1 <- pheatmap::pheatmap(score.matrix, cluster_rows = FALSE, silent = TRUE)
p2 <- pheatmap::pheatmap(score.matrix, scale = "column", cluster_rows = FALSE, silent = TRUE)
grid.arrange(p1[[4]],p2[[4]],nrow = 2) & NoLegend()#> NULL
同样使用基于correlation的两种计算方式,也都可以准确判定EC亚群的定位,并可以给出更准确的定位。
感兴趣的童鞋,可以测试一下用EE的Regional RNA-seq RPKM value预测EE细胞类群的定位!
计算不同预测方案的cluster * region 矩阵之间的相关性。
head(scRNAseq_Score_Compare(score.list,score.matrix),20)
#> UMI-20-Genes-40 UMI-200-Genes-40 UMI-2000-Genes-40 UMI-1500-Genes-50
#> 0.5777317 0.5777317 0.5777317 0.5716270
#> UMI-10-Genes-40 UMI-100-Genes-40 UMI-1000-Genes-40 UMI-10-Genes-50
#> 0.5565741 0.5565741 0.5565741 0.5516927
#> UMI-100-Genes-50 UMI-1000-Genes-50 UMI-20-Genes-20 UMI-200-Genes-20
#> 0.5516927 0.5516927 0.5444626 0.5444626
#> UMI-2000-Genes-20 UMI-10-Genes-100 UMI-100-Genes-100 UMI-1000-Genes-100
#> 0.5444626 0.5440390 0.5440390 0.5440390
#> UMI-1500-Genes-40 UMI-1500-Genes-100 UMI-10-Genes-30 UMI-100-Genes-30
#> 0.5420852 0.5397056 0.5365468 0.5365468更推荐的方案:从RNAseq的fastq文件入手,计算各个区域的高表达基因,做成基因集合,然后对每一个单细胞进行region定位的偏好性打分(AddModuleScore?),进而也能推断群水平的region偏好性。
pbmc <- AddModuleScore(scRNA, features = score.list$`20`$`20`$Genes)
DimPlot(pbmc, label = TRUE)FeaturePlot(pbmc,features = paste0("Cluster",1:5), ncol = 3)sessionInfo()
#> R version 4.3.0 (2023-04-21 ucrt)
#> Platform: x86_64-w64-mingw32/x64 (64-bit)
#> Running under: Windows 10 x64 (build 19045)
#>
#> Matrix products: default
#>
#>
#> locale:
#> [1] LC_COLLATE=Chinese (Simplified)_China.utf8
#> [2] LC_CTYPE=Chinese (Simplified)_China.utf8
#> [3] LC_MONETARY=Chinese (Simplified)_China.utf8
#> [4] LC_NUMERIC=C
#> [5] LC_TIME=Chinese (Simplified)_China.utf8
#>
#> time zone: Asia/Shanghai
#> tzcode source: internal
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ScoreRegionPreference_0.0.0.9000 gridExtra_2.3
#> [3] SeuratObject_4.1.3 Seurat_4.3.0.1
#>
#> loaded via a namespace (and not attached):
#> [1] deldir_1.0-9 pbapply_1.7-2 formatR_1.14
#> [4] rlang_1.1.1 magrittr_2.0.3 RcppAnnoy_0.0.21
#> [7] spatstat.geom_3.2-4 matrixStats_1.0.0 ggridges_0.5.4
#> [10] compiler_4.3.0 png_0.1-8 vctrs_0.6.2
#> [13] reshape2_1.4.4 stringr_1.5.0 pkgconfig_2.0.3
#> [16] fastmap_1.1.1 ellipsis_0.3.2 labeling_0.4.2
#> [19] utf8_1.2.3 promises_1.2.0.1 rmarkdown_2.22
#> [22] purrr_1.0.1 xfun_0.39 jsonlite_1.8.5
#> [25] goftest_1.2-3 highr_0.10 later_1.3.1
#> [28] spatstat.utils_3.0-3 irlba_2.3.5.1 parallel_4.3.0
#> [31] cluster_2.1.4 R6_2.5.1 ica_1.0-3
#> [34] stringi_1.7.12 RColorBrewer_1.1-3 spatstat.data_3.0-1
#> [37] reticulate_1.30 parallelly_1.36.0 lmtest_0.9-40
#> [40] scattermore_1.2 Rcpp_1.0.10 knitr_1.43
#> [43] tensor_1.5 future.apply_1.11.0 zoo_1.8-12
#> [46] VennDiagram_1.7.3 sctransform_0.3.5 httpuv_1.6.11
#> [49] Matrix_1.5-4 splines_4.3.0 igraph_1.4.3
#> [52] tidyselect_1.2.0 abind_1.4-5 rstudioapi_0.14
#> [55] yaml_2.3.7 spatstat.random_3.1-5 codetools_0.2-19
#> [58] miniUI_0.1.1.1 spatstat.explore_3.2-1 listenv_0.9.0
#> [61] lattice_0.21-8 tibble_3.2.1 plyr_1.8.8
#> [64] withr_2.5.0 shiny_1.7.4 ROCR_1.0-11
#> [67] evaluate_0.21 Rtsne_0.16 lambda.r_1.2.4
#> [70] future_1.33.0 survival_3.5-5 futile.logger_1.4.3
#> [73] polyclip_1.10-4 fitdistrplus_1.1-11 pillar_1.9.0
#> [76] KernSmooth_2.23-20 plotly_4.10.2 generics_0.1.3
#> [79] sp_2.0-0 ggplot2_3.4.4 munsell_0.5.0
#> [82] scales_1.3.0 globals_0.16.2 xtable_1.8-4
#> [85] glue_1.6.2 pheatmap_1.0.12 lazyeval_0.2.2
#> [88] tools_4.3.0 data.table_1.14.8 RANN_2.6.1
#> [91] leiden_0.4.3 cowplot_1.1.1 grid_4.3.0
#> [94] tidyr_1.3.0 colorspace_2.1-0 nlme_3.1-162
#> [97] patchwork_1.1.2 cli_3.6.1 spatstat.sparse_3.0-2
#> [100] futile.options_1.0.1 ineq_0.2-13 fansi_1.0.4
#> [103] viridisLite_0.4.2 dplyr_1.1.2 uwot_0.1.16
#> [106] gtable_0.3.3 digest_0.6.31 progressr_0.14.0
#> [109] ggrepel_0.9.3 farver_2.1.1 htmlwidgets_1.6.2
#> [112] htmltools_0.5.5 lifecycle_1.0.3 httr_1.4.6
#> [115] mime_0.12 MASS_7.3-58.4