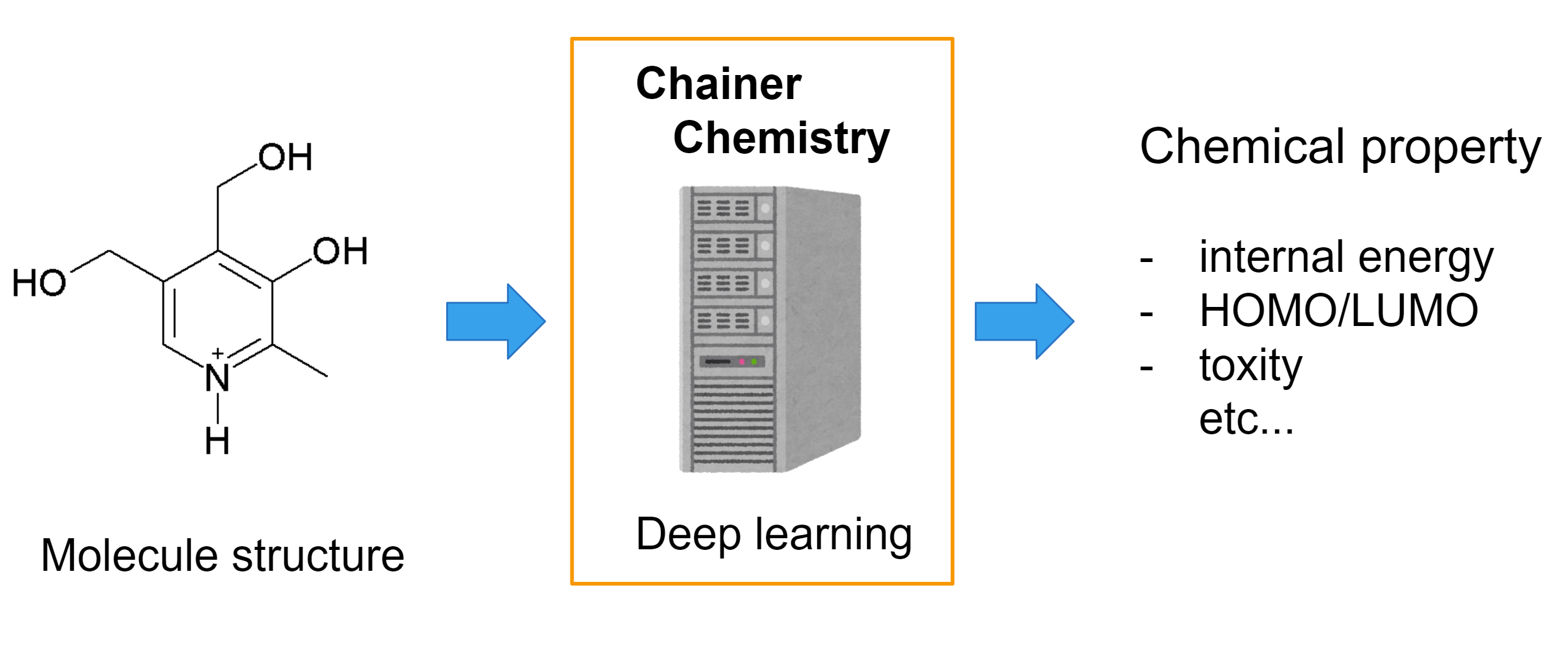
Chainer Chemistry: A Library for Deep Learning in Biology and Chemistry
Chainer Chemistry is a collection of tools to train and run neural networks for tasks in biology and chemistry using Chainer[1].
It supports various state-of-the-art deep learning neural network models (especially Graph Convolution Neural Network) for chemical molecule property prediction.
For more information, you can refer to documentation.
Quick start
1. Installation
Chainer Chemistry can be installed by pip command.
Note that it uses rdkit,
Open-Source Cheminformatics Software.
Below code is an example to install rdkit by conda command provided by
anaconda.
pip install chainer-chemistry
conda install -c rdkit rdkit2. Run example training code
The official repository provides examples several graph convolution networks with the Tox21 and QM9 datasets (the Tox21 example has inference code as well). You can obtain the code by cloning the repository:
git clone https://github.com/pfnet-research/chainer-chemistry.gitThe following code is how to train Neural Fingerprint (NFP) with the Tox21 dataset on CPU:
cd chainer-chemistry/examples/tox21
python train_tox21.py --method=nfp --gpu=-1 # set --gpu=0 if you have GPU
Installation
Usual users can install this library via PyPI:
pip install chainer-chemistry
Chainer Chemistry is still in experimental development. If you would like to use latest sources. please install master branch with the command:
git clone https://github.com/pfnet-research/chainer-chemistry.git
pip install -e chainer-chemistry
Dependencies
Following packages are required to install Chainer Chemistry and are automatically
installed when you install it by pip command.
Also, it uses following library, you need to manually install it.
See the official document
for installation.
If you have setup anaconda, you may install rdkit by following command.
conda install -c rdkit rdkit
Supported model
Currently, following graph convolutional neural networks are implemented.
- NFP: Neural fingerprint [2, 3]
- GGNN: Gated Graph Neural Network [4, 3]
- Weave: [5, 3]
- SchNet: [6]
Supported dataset
Currently, following dataset is supported.
- QM9 [7, 8]
- Tox21 [9]
License
MIT License.
We provide no warranty or support for this implementation. Each model performance is not guaranteed, and may not achieve the score reported in each paper. Use it at your own risk.
Please see the LICENSE file for details.
Links
Links for Chainer Chemistry:
- Document: https://chainer-chemistry.readthedocs.io
- Blog: Release Chainer Chemistry: A library for Deep Learning in Biology and Chemistry
Links for other Chainer projects:
- Chainer: A flexible framework of neural networks for deep learning
- Official page: Website
- Github: chainer/chainer
- ChainerRL: Deep reinforcement learning library built on top of Chainer - chainer/chainerrl
- ChainerCV: A Library for Deep Learning in Computer Vision - chainer/chainercv
- ChainerMN: Scalable distributed deep learning with Chainer - chainer/chainermn
- ChainerUI: User Interface for Chainer - chainer/chainerui
- PaintsChainer: Line drawing colorization using chainer
- Official page: Website
- Github: pfnet/PaintsChainer
- CuPy: NumPy-like API accelerated with CUDA
If you are new to chainer, here is a tutorial to start with:
- Chainer Notebooks: hands on tutorial - mitmul/chainer-handson
Reference
[1] Seiya Tokui, Kenta Oono, Shohei Hido, and Justin Clayton. Chainer: a next-generation open source framework for deep learning. In Proceedings of Workshop on Machine Learning Systems (LearningSys) in Advances in Neural Information Processing System (NIPS) 28, 2015.
[2] David K Duvenaud, Dougal Maclaurin, Jorge Iparraguirre, Rafael Bombarell, Timothy Hirzel, Alan Aspuru-Guzik, and Ryan P Adams. Convolutional networks on graphs for learning molecular fingerprints. In C. Cortes, N. D. Lawrence, D. D. Lee, M. Sugiyama, and R. Garnett, editors, Advances in Neural Information Processing Systems (NIPS) 28, pages 2224–2232. Curran Asso- ciates, Inc., 2015.
[3] Justin Gilmer, Samuel S Schoenholz, Patrick F Riley, Oriol Vinyals, and George E Dahl. Neural message passing for quantum chemistry. arXiv preprint arXiv:1704.01212, 2017.
[4] Yujia Li, Daniel Tarlow, Marc Brockschmidt, and Richard Zemel. Gated graph sequence neural networks. arXiv preprint arXiv:1511.05493, 2015.
[5] Steven Kearnes, Kevin McCloskey, Marc Berndl, Vijay Pande, and Patrick Riley. Molecular graph convolutions: moving beyond fingerprints. Journal of computer-aided molecular design, 30(8):595–608, 2016.
[6] Kristof Schütt, Pieter-Jan Kindermans, Huziel Enoc Sauceda Felix, Stefan Chmiela, Alexandre Tkatchenko, and Klaus-Rober Müller Schnet: A continuous-filter convolutional neural network for mod eling quantum interactions. In I. Guyon, U. V. Luxburg, S. Bengio, H. Wallach, R. Fergus, S. Vishwanathan, and R. Garnett, editors, Advances in Neural Information Processing Systems (NIPS) 30, pages 992–1002. Curran Associates, Inc., 2017.
[7] Lars Ruddigkeit, Ruud Van Deursen, Lorenz C Blum, and Jean-Louis Reymond. Enumeration of 166 billion organic small molecules in the chemical universe database gdb-17. Journal of chemical information and modeling, 52(11):2864–2875, 2012.
[8] Raghunathan Ramakrishnan, Pavlo O Dral, Matthias Rupp, and O Anatole Von Lilienfeld. Quantum chemistry structures and properties of 134 kilo molecules. Scientific data, 1:140022, 2014.
[9] Ruili Huang, Menghang Xia, Dac-Trung Nguyen, Tongan Zhao, Srilatha Sakamuru, Jinghua Zhao, Sampada A Shahane, Anna Rossoshek, and Anton Simeonov. Tox21challenge to build predictive models of nuclear receptor and stress response pathways as mediated by exposure to environmental chemicals and drugs. Frontiers in Environmental Science, 3:85, 2016.