The code uses mixtures of state space models to perform unsupervised clustering of short trajectories. Within the state space framework, we let expensive-to-gather biomarkers correspond to hidden states and readily obtainable cognitive metrics correspond to measurements. Upon training with expectation maximization, we often find that our clusters stratify persons according to clinical outcome. Furthermore, we can effectively predict on held-out trajectories using cognitive metrics alone. Our approach accommodates missing data through model marginalization and generalizes across research and clinical cohorts.
We consider a training dataset
consisting of np.nan's to shorter trajectories.
We adopt a mixture of state space models for the data:
Each individual
In our main framework, we additionally assume that the cluster-specific state
initialisation is Gaussian, i.e.
In particular, we assume that the variables we are modeling are continuous and
changing over time. When we train a model like the above, we take a dataset
- (E) Expectation step: given the current model, we assign each data instance
$(z^i_{1:T}, x^i_{1:T})$ to the cluster to which it is mostly likely to belong under the current model - (M) Maximization step: given the current cluster assignments, we compute the
sample-level cluster assignment probabilities (the
$\pi_c$ ) and optimal cluster-specific parameters
Optimization completes after a fixed (large) number of steps or when no data instances change their cluster assignment at a given iteration.
All model training and predictions are done in Python. We provide a pip-installable package unsupervised-multimodal-trajectory-modeling to do much of the work for you. It can be installed with dependencies as follows:
pip3 install -r requirements.txt
or in a virtual environment with:
python3 -m venv venv
source venv/bin/activate
pip3 install -r requirements.txt
A typical workflow for a new dataset looks like this:
-
Create a new
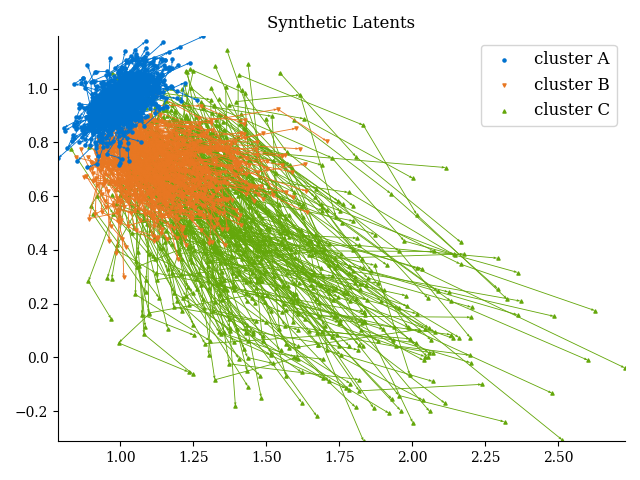
data_*.pyfile that organises the data into tensors of states and measurements as described above. Often, we include functionality for plotting trajectories of states by inferred cluster here. This is also a useful place to wrangle auxiliary data that will then be used to profile learned clusters. For example data_synthetic.py, gives us trajectories in state space that look like this: -
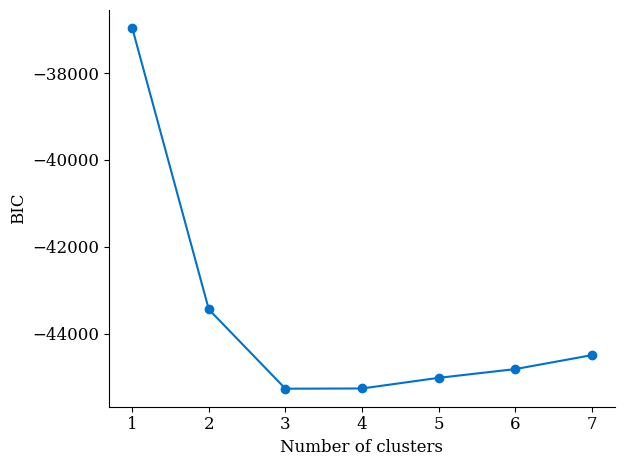
Run a model selection script to create plots of BIC vs. number of clusters. Import the
data_*.pyfile you created in the previous step and modify model_selection_synthetic.py as needed. This will give us a plot like so:We know the data was generated from a 3-component mixture model, and the BIC plot above reflects that. We now focus on models with 3 clusters.
-
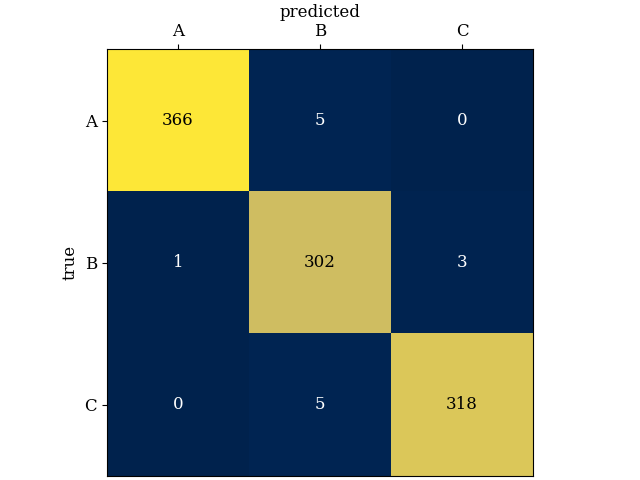
Experiment with training and testing, like in inference_synthetic.py. We can, for example, train a model on one set and then predict cluster membership for a new dataset. When both datasets were generated according to the same specifications, we find very good agreement for this example, even when predictions are made without access to the states. Here's a confusion matrix (having generated the test set, we know ground truth):
For real-life examples, we are often interested in profiling our clusters to better understand them.
-
We have an extended framework to incorporate nonlinearities into our state and measurement models. Check out inference_nonlinear.py for a minimal example.
Some efforts have been made to handle edge cases. For a given training run, if any cluster becomes too small, training automatically terminates.
Footnotes
-
A. Dempster, N. Laird, and D. B. Rubin. Maximum Likelihood from
Incomplete Data via the EM Algorithm. J. Roy. Stat. Soc. Ser. B (Stat. Methodol.) 39.1 (1977), pp. 1–38. ↩