Fabio Votta 29 August 2018
generate_multi <- function(n, cor_seq){
set.seed(2017)
x <- runif(n, 1, 10)
models <- list()
std.models <- list()
for (jj in seq_along(cor_seq)) {
## generate correlated variable x2
dat <- data.frame(corgen(x = x, r = cor_seq[jj], epsilon = 0))
colnames(dat) <- c("x1", "x2")
## generate y variable
dat$y <- 0.5 * dat$x1 + 0.5 * dat$x2 + rnorm(n, sd = 10)
## modelling and tidy dataframe
models[[jj]] <- tidy(lm(y ~ x1 + x2, data = dat))
## get standardized betas
std.models[[jj]] <- data.frame(lm.beta::coef.lm.beta((lm.beta::lm.beta(lm(y ~ x1 + x2, data = dat)))))
colnames(std.models[[jj]]) <- c("std.estimate")
## bind it together
models[[jj]] <- std.models[[jj]] %>%
bind_cols(models[[jj]])
models[[jj]]$cors <- cor_seq[jj]
}
sim_dat <- bind_rows(models)
sim_dat$col <- n
return(sim_dat)
}
draw.data <- function(cor_seq = NULL, step_seq = NULL){
sim.list <- list()
for(jj in seq_along(step_seq)) {
sim.list[[jj]] <- generate_multi(n = step_seq[jj], cor_seq)
sim.list[[jj]]$n <- step_seq[jj]
cat(paste0("Batch: ", jj, "\t"))
}
sim_data <- bind_rows(sim.list)
return(sim_data)
}sim_data <- draw.data(cor_seq = seq(0,.99,0.01), step_seq = seq(50, 10000, by = 50))
if(!dir.exists("data")) dir.create("data")
save(sim_data, file = "data/sim_data.Rdata")load("data/sim_data.Rdata")get_smooths <- function(smooth_dat, n_val, y) {
smooth_dat <- filter(smooth_dat, n == n_val & term == "x1")
# y <- enquo(y)
fm <- paste0(y," ~ cors")
smooth_vals <- predict(loess(fm, smooth_dat), smooth_dat$cors)
smooth_dat %>%
mutate(smooth = smooth_vals) %>%
group_by(n) %>%
summarise(max_smooth = max(smooth),
min_smooth = min(smooth)
)
}
smooth_dat <- c(50, 100, 150, 200, 10000) %>%
map_df(~get_smooths(sim_data, n_val = .x, y = "std.error")) %>%
mutate(n_lab = ifelse(n == 50, "Sample Size: 50", n))
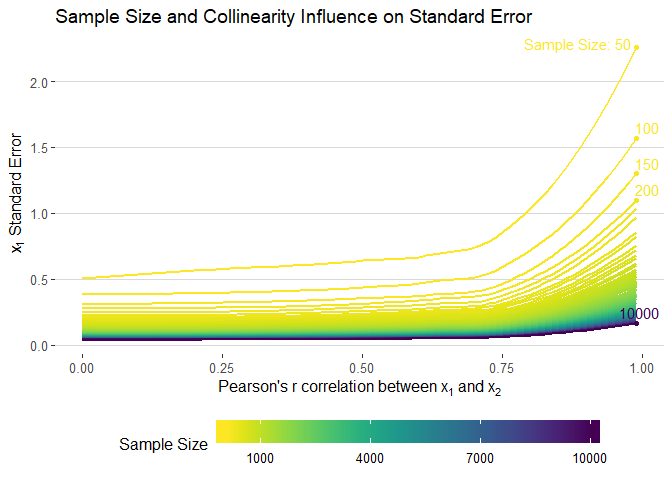
sim_data %>%
filter(term == "x1") %>%
ggplot(aes(cors, std.error, colour = n, group = n)) +
geom_smooth(method = "loess", se = F, size = 1, alpha = 0.5) +
xlab(expression("Pearson's"~r~correlation~between~x[1]~and~x[2])) +
ylab(expression(x[1]~Standard~Error)) +
theme_hc() +
scale_color_viridis("Sample Size", direction = -1,
# limits = seq(1000, 10000, 3000),
breaks = seq(1000, 10000, 3000),
labels = seq(1000, 10000, 3000)) +
ggtitle("Sample Size and Collinearity Influence on Standard Error") +
geom_point(data = smooth_dat, aes(x = .99, y = max_smooth)) +
geom_text_repel(data = smooth_dat, aes(x = .99, y = max_smooth, label = n_lab),
nudge_y = 0.07, nudge_x = 0.03) +
guides(colour = guide_colourbar(barwidth = 20, label.position = "bottom"))ggsave(filename = "images/std_static.png", width = 10, height = 7)sim_data %>%
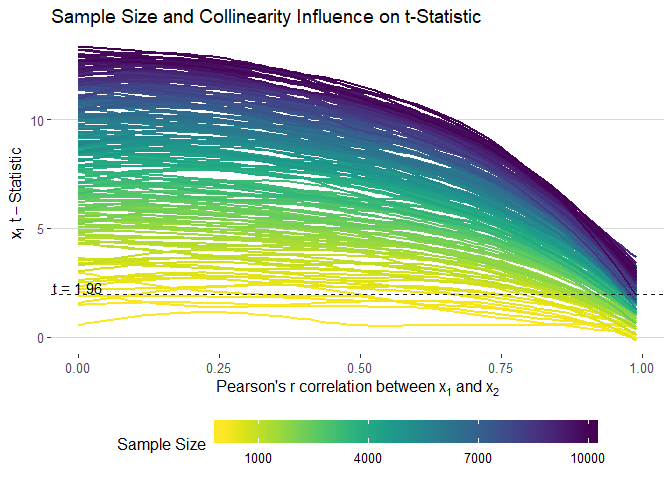
filter(term == "x1") %>%
ggplot(aes(cors, statistic, colour = n, group = n)) +
geom_smooth(method = "loess", se = F, size = 1, alpha = 0.5) +
xlab(expression("Pearson's"~r~correlation~between~x[1]~and~x[2])) +
ylab(expression(x[1]~t-Statistic)) +
theme_hc() +
scale_color_viridis("Sample Size", direction = -1,
# limits = seq(1000, 10000, 3000),
breaks = seq(1000, 10000, 3000),
labels = seq(1000, 10000, 3000)) +
geom_hline(yintercept = 1.96, linetype = "dashed", alpha = 0.9) +
annotate(geom = "text", x = 0, y = 2.3, label = "t = 1.96") +
ggtitle("Sample Size and Collinearity Influence on t-Statistic") +
guides(colour = guide_colourbar(barwidth = 20, label.position = "bottom"))ggsave(filename = "images/t_static.png", width = 10, height = 7)sim_data %>%
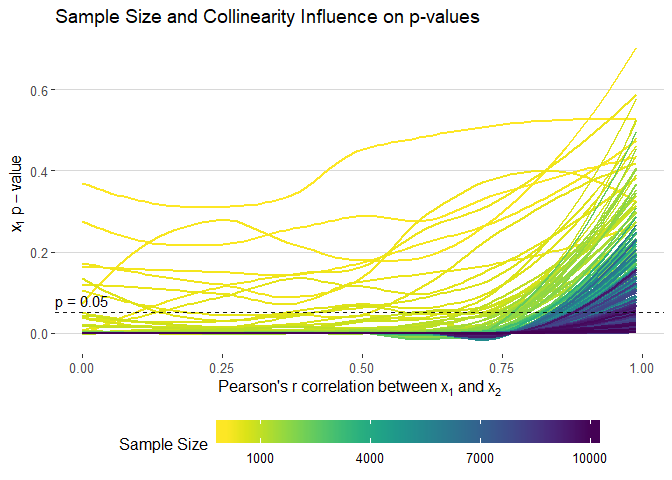
filter(term=="x1") %>%
ggplot(aes(cors, p.value, colour = n, group = n)) +
geom_smooth(method = "loess", se = F, size = 1, alpha = 0.5) +
xlab(expression("Pearson's"~r~correlation~between~x[1]~and~x[2])) +
ylab(expression(x[1]~p-value)) +
theme_hc() +
scale_color_viridis("Sample Size", direction = -1,
# limits = seq(1000, 10000, 3000),
breaks = seq(1000, 10000, 3000),
labels = seq(1000, 10000, 3000)) +
geom_hline(yintercept = 0.05, linetype = "dashed", alpha = 0.9) +
annotate(geom = "text", x = 0, y = 0.08, label = "p = 0.05") +
ggtitle("Sample Size and Collinearity Influence on p-values") +
guides(colour = guide_colourbar(barwidth = 20, label.position = "bottom"))ggsave(filename = "images/p_static.png", width = 10, height = 7)sim_data %>%
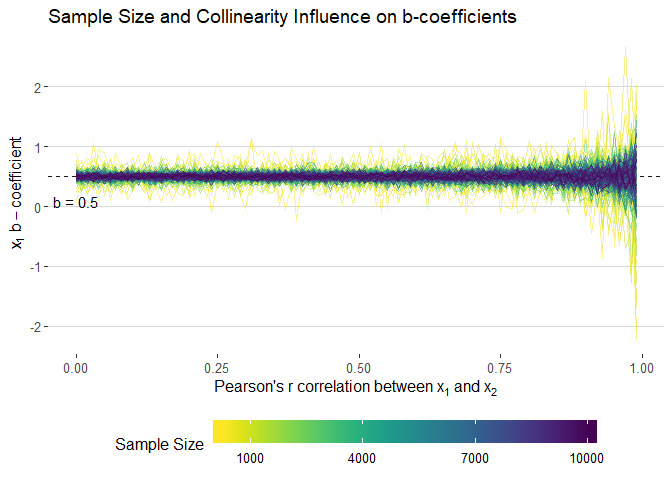
filter(term=="x1") %>%
filter(n>200) %>%
ggplot(aes(cors, estimate, colour = n, group = n)) +
geom_hline(yintercept = 0.5, linetype = "dashed", alpha = 0.9) +
#geom_smooth(method = "loess", se = F, size = 1, alpha = 0.5) +#
geom_line(alpha = 0.5) +
xlab(expression("Pearson's"~r~correlation~between~x[1]~and~x[2])) +
ylab(expression(x[1]~b-coefficient)) +
theme_hc() +
scale_color_viridis("Sample Size", direction = -1,
# limits = seq(1000, 10000, 3000),
breaks = seq(1000, 10000, 3000),
labels = seq(1000, 10000, 3000)) +
annotate(geom = "text", x = 0, y = 0.08, label = "b = 0.5") +
ggtitle("Sample Size and Collinearity Influence on b-coefficients") +
guides(colour = guide_colourbar(barwidth = 20, label.position = "bottom"))ggsave(filename = "images/b_static.png", width = 10, height = 7)library(gganimate)
sim_data_sub <- sim_data %>%
filter(term == "x1") %>%
filter(n > 200) %>%
filter(n %in% c(300, 1000, 10000)) %>%
mutate(estimate_lab = round(estimate, 2) %>% as.character) %>%
mutate(n = as.factor(n))
anim1 <- sim_data_sub %>%
# filter(term == "x1") %>%
# filter(n > 200) %>%
ggplot(aes(cors, estimate, colour = n, group = n)) +
geom_hline(yintercept = 0.5, linetype = "dashed", alpha = 0.9) +
geom_line() +
geom_segment(aes(xend = 1, yend = estimate), linetype = 2, colour = 'grey') +
geom_point(size = 2) +
geom_text(aes(x = 1, label = n), hjust = 0, size = 4, fontface = "bold") +
geom_text(aes(x = 0.15, y = 1.8, label = paste0("Correlation: ", cors)),
hjust = 1, size = 5, color = "black") +
geom_text(aes(label = estimate_lab), hjust = 0, size = 3, fontface = "bold", nudge_y = 0.1) +
xlab("Pearson's r correlation between x1 and x2") +
ylab("x1 b-coefficient") +
coord_cartesian(clip = 'off') +
theme_hc() +
scale_color_viridis("Sample Size",
direction = -1,
discrete = T,
begin = 0.3,
# limits = seq(1000, 10000, 3000),
breaks = seq(1000, 10000, 3000),
labels = seq(1000, 10000, 3000)) +
guides(colour = F) +
theme(title = element_text(size = 15, face = "bold"),
axis.text.x = element_text(size = 14, face = "bold"),
axis.text.y = element_text(size = 10, face = "italic")) +
ggtitle("Sample Size and Collinearity Influence on b-coefficients (Sample Sizes: 300, 1000 and 10.000)") +
# guides(colour = guide_colourbar(barwidth = 20, label.position = "bottom")) +
# Here comes the gganimate code
transition_reveal(n, cors)
anim1 %>% animate(
nframes = 500, fps = 15, width = 1000, height = 600, detail = 3
)
anim_save("images/b_anim.gif")sim_data %>%
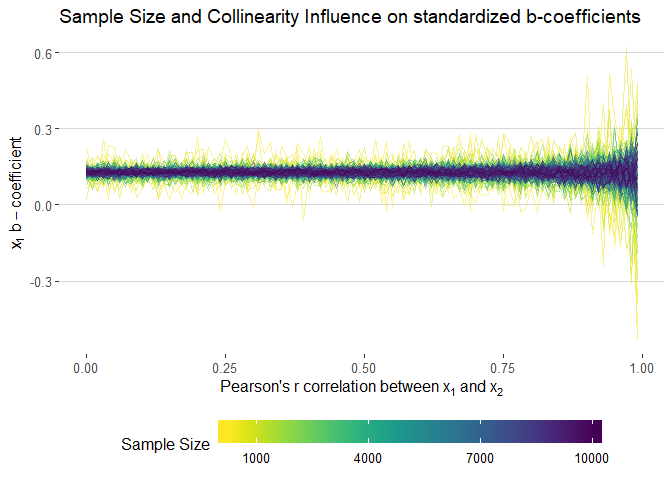
filter(term=="x1") %>%
filter(n>200) %>%
ggplot(aes(cors, std.estimate, colour = n, group = n)) +
#geom_smooth(method = "loess", se = F, size = 1, alpha = 0.5) +
geom_line(alpha = 0.5) +
xlab(expression("Pearson's"~r~correlation~between~x[1]~and~x[2])) +
ylab(expression(x[1]~b-coefficient)) +
theme_hc() +
scale_color_viridis("Sample Size", direction = -1,
# limits = seq(1000, 10000, 3000),
breaks = seq(1000, 10000, 3000),
labels = seq(1000, 10000, 3000)) +
ggtitle("Sample Size and Collinearity Influence on standardized b-coefficients") +
guides(colour = guide_colourbar(barwidth = 20, label.position = "bottom"))ggsave(filename = "images/b_standardized_static.png", width = 10, height = 7)sessionInfo()## R version 3.5.0 (2018-04-23)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17134)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=German_Germany.1252 LC_CTYPE=German_Germany.1252
## [3] LC_MONETARY=German_Germany.1252 LC_NUMERIC=C
## [5] LC_TIME=German_Germany.1252
##
## attached base packages:
## [1] grid stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] bindrcpp_0.2.2 ggrepel_0.8.0 lm.beta_1.5-1
## [4] gridExtra_2.3 viridis_0.5.1 viridisLite_0.3.0
## [7] ecodist_2.0.1 forcats_0.3.0 stringr_1.3.0
## [10] dplyr_0.7.5 readr_1.1.1 tidyr_0.8.1
## [13] tibble_1.4.2 ggplot2_3.0.0.9000 tidyverse_1.2.1
## [16] ggthemes_4.0.0 broom_0.4.4 purrr_0.2.4
## [19] arm_1.10-1 lme4_1.1-17 Matrix_1.2-14
## [22] MASS_7.3-49
##
## loaded via a namespace (and not attached):
## [1] Rcpp_0.12.18 lubridate_1.7.4 lattice_0.20-35 assertthat_0.2.0
## [5] rprojroot_1.3-2 digest_0.6.15 psych_1.8.3.3 R6_2.2.2
## [9] cellranger_1.1.0 plyr_1.8.4 backports_1.1.2 evaluate_0.10.1
## [13] coda_0.19-1 httr_1.3.1 pillar_1.2.1 rlang_0.2.1
## [17] lazyeval_0.2.1 readxl_1.1.0 minqa_1.2.4 rstudioapi_0.7
## [21] nloptr_1.0.4 rmarkdown_1.9 labeling_0.3 splines_3.5.0
## [25] foreign_0.8-70 munsell_0.4.3 compiler_3.5.0 modelr_0.1.1
## [29] pkgconfig_2.0.1 mnormt_1.5-5 htmltools_0.3.6 tidyselect_0.2.4
## [33] crayon_1.3.4 withr_2.1.2 nlme_3.1-137 jsonlite_1.5
## [37] gtable_0.2.0 pacman_0.4.6 magrittr_1.5 scales_0.5.0
## [41] cli_1.0.0 stringi_1.1.7 reshape2_1.4.3 xml2_1.2.0
## [45] tools_3.5.0 glue_1.3.0 hms_0.4.2 abind_1.4-5
## [49] parallel_3.5.0 yaml_2.1.19 colorspace_1.4-0 rvest_0.3.2
## [53] knitr_1.20 bindr_0.1.1 haven_1.1.2