Knights et al. (2023). Neural Evidence of Functional Compensation for Fluid Intelligence in Healthy Ageing.
This repository contains code accompanying the multivariate Bayesian machine-learning project described in the following preprint:
Knights, E., Henson, R. N., Morcom, A. M., Mitchell, D. J., & Tsvetanov, K. A. (2023). Neural Evidence of Functional Compensation for Fluid Intelligence in Healthy Ageing. bioRxiv, 2023-09.
Modelling Quickstart
Clone & download processed fMRI/behavioural data with shell from root:
git clone https://github.com/ethanknights/Knightsetal_fMRI-Cattell-Compensation
cd Knightsetal_fMRI-Cattell-Compensation
./setup.shUse R to perform univariate/multivariate modelling analysis (see README sections 2/4 in 'Analysis Pipline') e.g.:
setwd('R')
source('load_data.R')
source('plot_ROI.R') #For interaction: activation ~ age * bhvPrerequisites
Software
- FSL v4.0+
- Matlab (tested with r2019a)
- SPM12
https://www.fil.ion.ucl.ac.uk/spm/software/spm12/ - Commonality Analysis tools
https://github.com/kamentsvetanov/CommonalityAnalysis - BrainNet toolbox
https://www.nitrc.org/projects/bnv/
- SPM12
- R
- MASS
- BayesFactor
- ggeffects
- Popular manipulation/visualisation packages (e.g., ggplot2, dplyr, tidyverse)
- Python
- gl package (to plot t-statistic maps in mricroGL)
Datasets
Raw data for Machine-Learning
- For performing Multivariate Bayes and the preceding whole-brain univariate analysis (e.g. generating second-level models, defining ROIs, extracting timeseries), download the Cattell first-level model images derived from automatic analysis (see Taylor et al. 2017 NeuroImage) by submitting a Cam-CAN data-request:
https://camcan-archive.mrc-cbu.cam.ac.uk/dataaccess
Summary Data for Modelling
- For hypothesis testing using R download data from the Open Science Framework (OSF).
Linux/Mac
./setup.sh #Download, extract & move the summary datasets from the OSF, to their required destinationsWindows
- Manaully download/extract the summary_data.zip from
https://osf.io/v7kmh/and move each.csvto the following corresponding analysis directories:
Univariate
mkdir R/csv
mv univariate.csv R/csv/Univariate (Vascular control)
mkdir R_vascular/csv
mv univariate_vascular_RSFA.csv R_vascular/csv/MVB
# Cuneal Cortex
mkdir MVB/R/dropMVBSubjects-1/70voxel_model-sparse/csv
mv MVB_cuneal.csv MVB/R/dropMVBSubjects-1/70voxel_model-sparse/csv/
# Frontal cortex
mkdir MVB/R/dropMVBSubjects-1/control-frontalROI_70voxel_model-sparse/csv
mv MVB_frontal.csv MVB/R/dropMVBSubjects-1/control-frontalROI_70voxel_model-sparse/csvMVB (Multivariate mapping)
# Cuneal Cortex
mv MVB_multvariateMapping-Cuneal/extradata_ShuffledGroupFVals.csv MVB/R/dropMVBSubjects-1/70voxel_model-sparse/csv/
# Frontal cortex
mv MVB_multvariateMapping-Frontal/extradata_ShuffledGroupFVals.csv MVB/R/dropMVBSubjects-1/control-frontalROI_70voxel_model-sparse/csvAnalysis Pipeline
1) Whole-brain Univariate Analysis
Use matlab to run fMRI preprocessing and generate the group univariate contrasts (i.e., second level model generation, TFCE correction, ROI definition and timeseries extraction):
pipeline.m
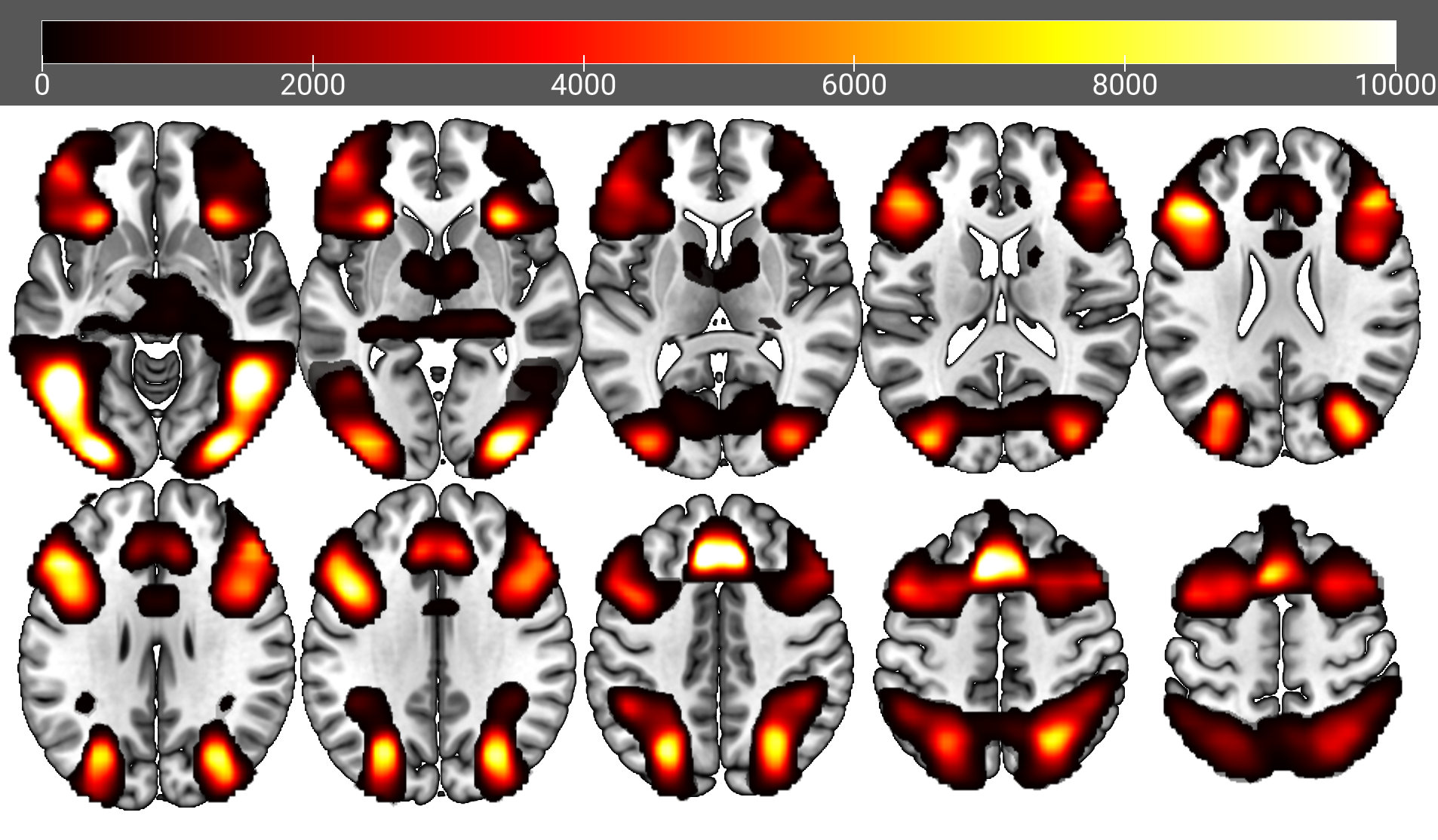
/* Alternatively, download the summary datasets described earlier in the Prerequisties section.Use Python to plot TFCE-corrected t-statistic maps with MRIcroGL.
./python/mosaic_MRICRON_HardBIGEasy.py
gl.overlayload('./nii_HardBIGEasy/Intercept_tfce197.nii')
gl.minmax(2, 0, 10000)
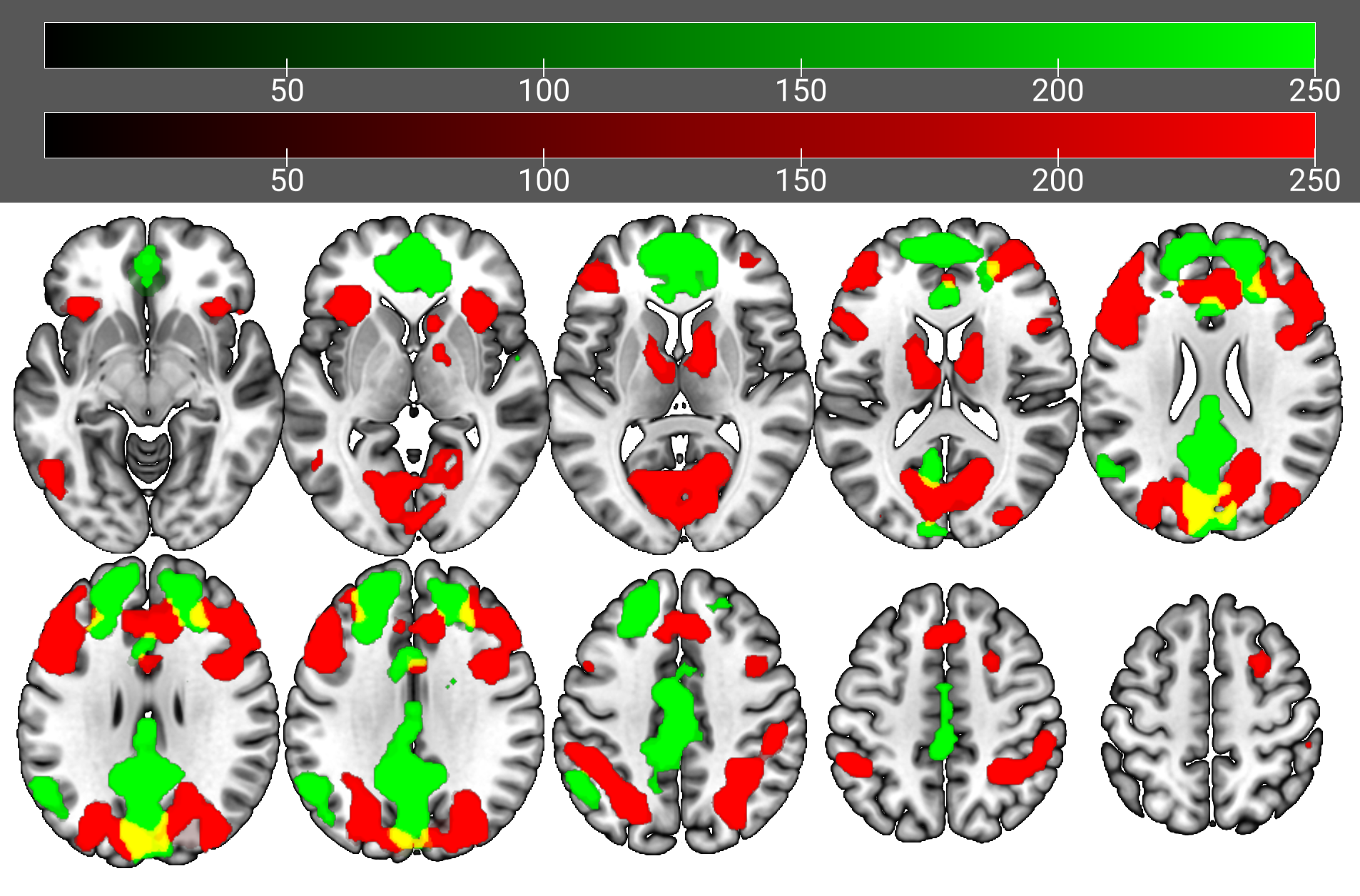
gl.colorname (1,"1red")./python/mosaic_MRICRON_Covariates.py:
gl.overlayload('/nii_Covariates/tval_Age_tfce197.nii')
gl.minmax(1, 0, 250)
gl.colorname (1,"2green")
gl.overlayload('nii_Covariates/tval_PC6_tfce197.nii')
gl.minmax(2, 0, 250)
gl.colorname (2,"1red")2) Univariate Modelling
In R, generate the regression models & plots:
setwd('R')
source('loadData.R')
#For interaction: Bhv ~ age * gender
source('plot_bhv.R')
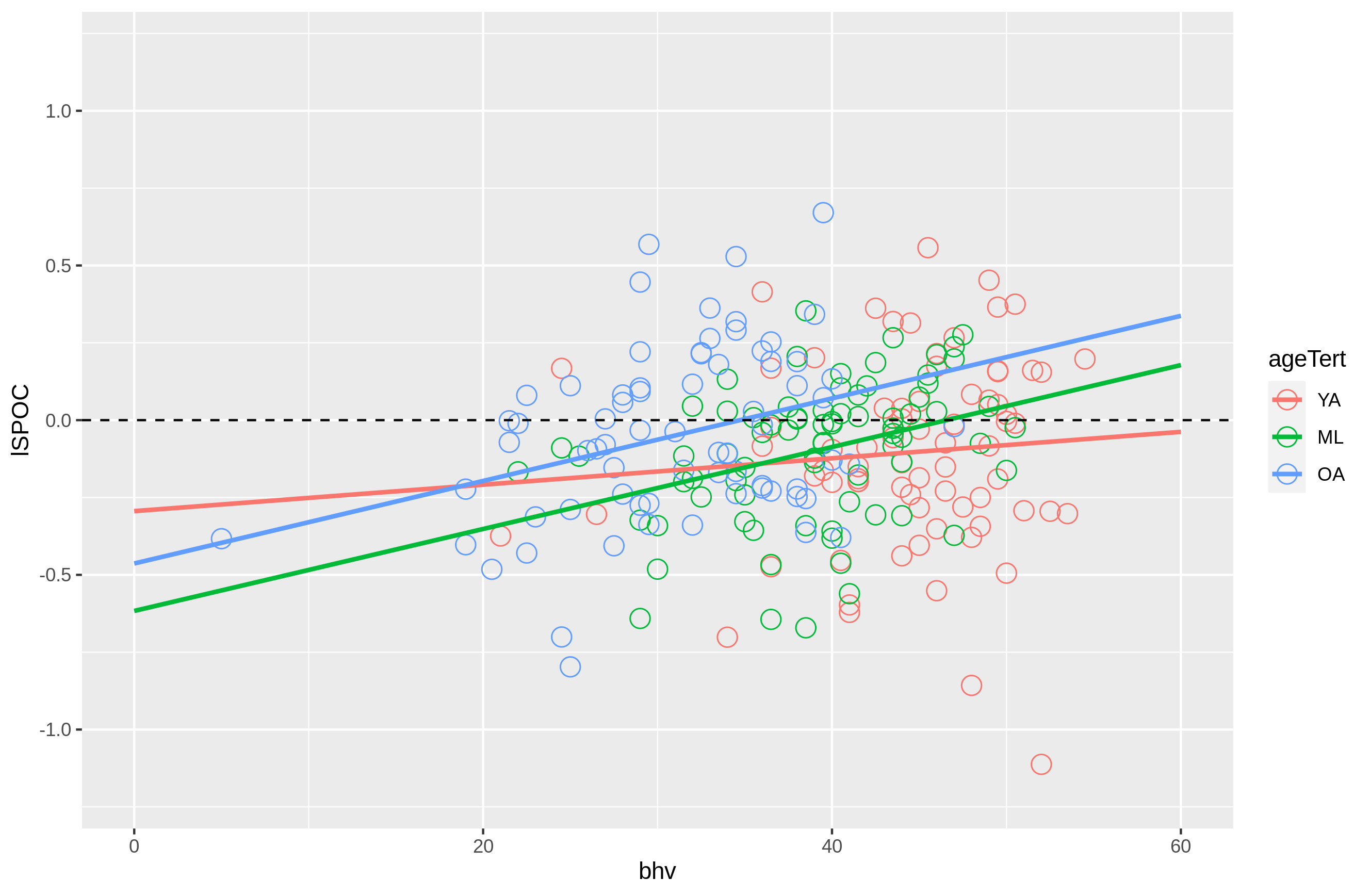
#For interaction: Activation ~ age * bhv
source('plot_ROI.R')
#For the control vascular RSFA analysis:
setwd('R_vascular')
source('load_data_RSFA.R')
source('plot_ROI_RSFA.R')3) Multivariate Bayes
Use matlab to run the MVB machine-learning pipeline:
cd MVB
setupDir.m
wrapper.m
doPostProcessing.m
/* Alternatively, download the summary datasets described earlier in the Prerequisties section.4) Multivariate Modelling
In R, generate the logistic regression models & plots for the ROI datasets independently:
#CHOOSE ONE ROI:
setwd('./MVB/R/dropMVBSubjects-1/70voxel_model-spare') #Cuneal Cortex
setwd('./MVB/R/dropMVBSubjects-1/control_frontalROI_70voxel_model-spare') #Frontal Cortex
#Load data into dataframe 'df'
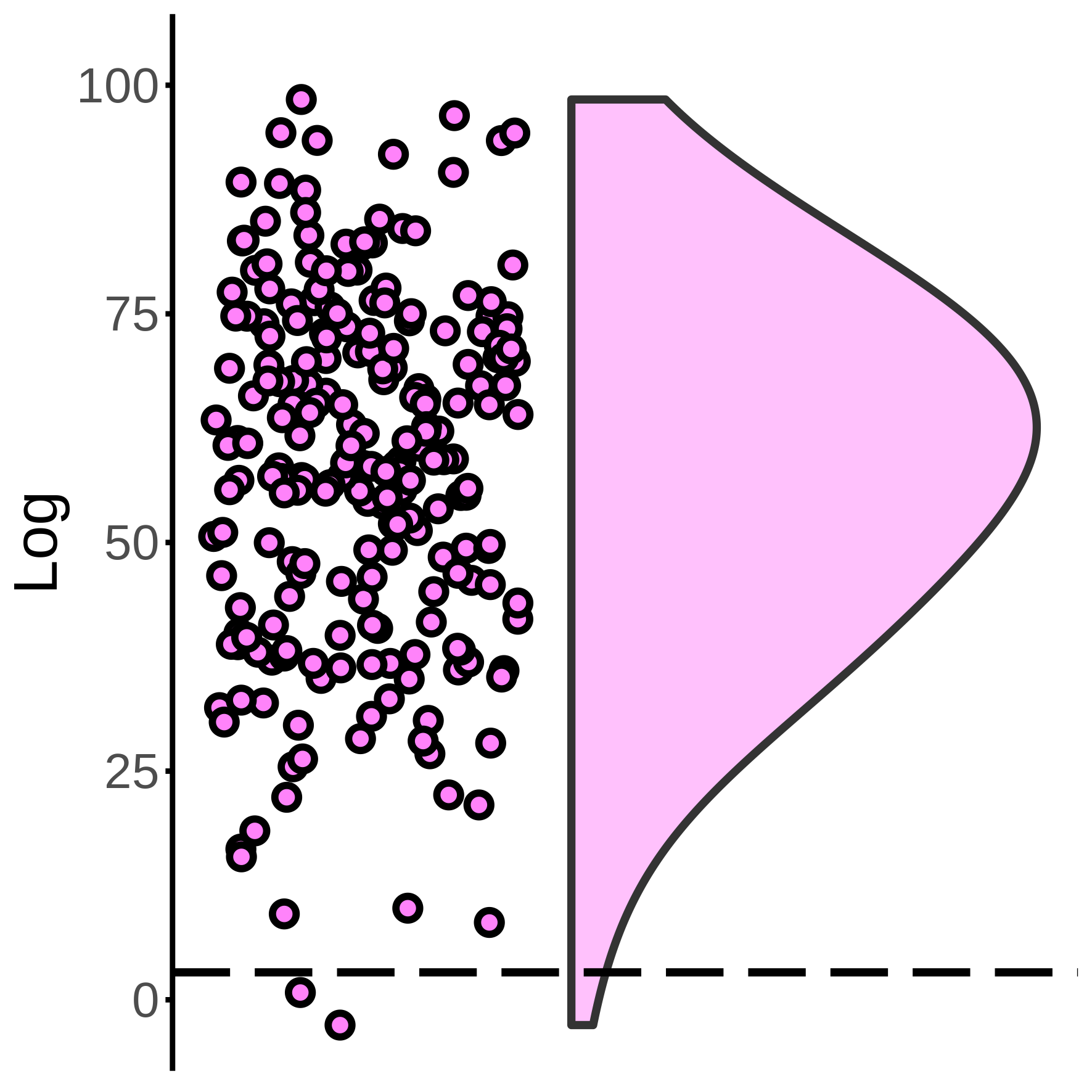
source('run_loadData.R')For testing the multvariate mapping, use raincloud package:
source('run_MVB_shuffledMapping.R') For logistic regression of Boost likelihood, use MASS/polr packages:
source('run_fMRI_MVB.R')