A suite of software tools to analyze nucleosome positioning sequence patterns. These patterns are represented by distributions of frequencies of dinucleotide occurrences in a stack of DNA sequences that were bound by nucleosomes.
Motivation to write these utilities was that most often, computations of such patterns are implemented by researchers individually. This makes it difficult to reproduce the results obtained in different projects and to perform comparisons.
The dnpatterntools consist of utlities to convert fasta sequences into binary strings of dinucleotide occurrences, to compute dinucleotide frequencies of occurrence in a batch of aligned fasta sequences, to compute correlations between patterns of dinucleotide distributions on forward and complementary sequences bound by nucleosomes, to smooth the patterns and to compute their periodogramms.
The dnpatterntools can be used as stanalone programs or Galay tools. The dockerized galaxy instance here runs on any local machine with the docker installed. This instance is based on the galaxy-stable . Working dnapatterntools utilities can be found on Galaxy Test Tool Shed. Here, the fully functional Galaxy wrappers are provided in the tools folder. To try a dnpatterntools Galaxy instance on your Linux machine clone the dnpatterntools, cd to the tools folder and run a planemo serve from within ~/dnpatterntools/tools folder. It will launch a ready to use dnpatterntools Galaxy instance.
How to use tools in Galaxy is explained here.
Core utilities are written in C++ using the SeqAn library and can be installed on Linux system by conda:
conda install dnp-binstrings -c bioconda conda install dnp-diprofile -c bioconda conda install dnp-corrprofile -c bioconda conda install dnp-fourier -c bioconda
To build core utilities from source:
cd <your working dir> git clone https://github.com/erinijapranckeviciene/dnpatterntools.git cd dnpatterntools
and follow instructions .
Complete dnpaterntools workflow has following steps:
- Computation of dinucleotide frequency distributions in a batch of aligned sequences.
- Determining a nucleosome location from the dinucleotide frequency distributions.
- Symmetrization and computation of composite distributions of WW/SS (W = A or T and S=C or G) and RR/YY (R=A or G and Y=C or T) dinucleotides.
- Normalization and smoothing of the dinucleotide frequency distribution patterns in nucleosomes and computing their periodograms.
Workflow steps and tools required in each step are shown in Figure 1.
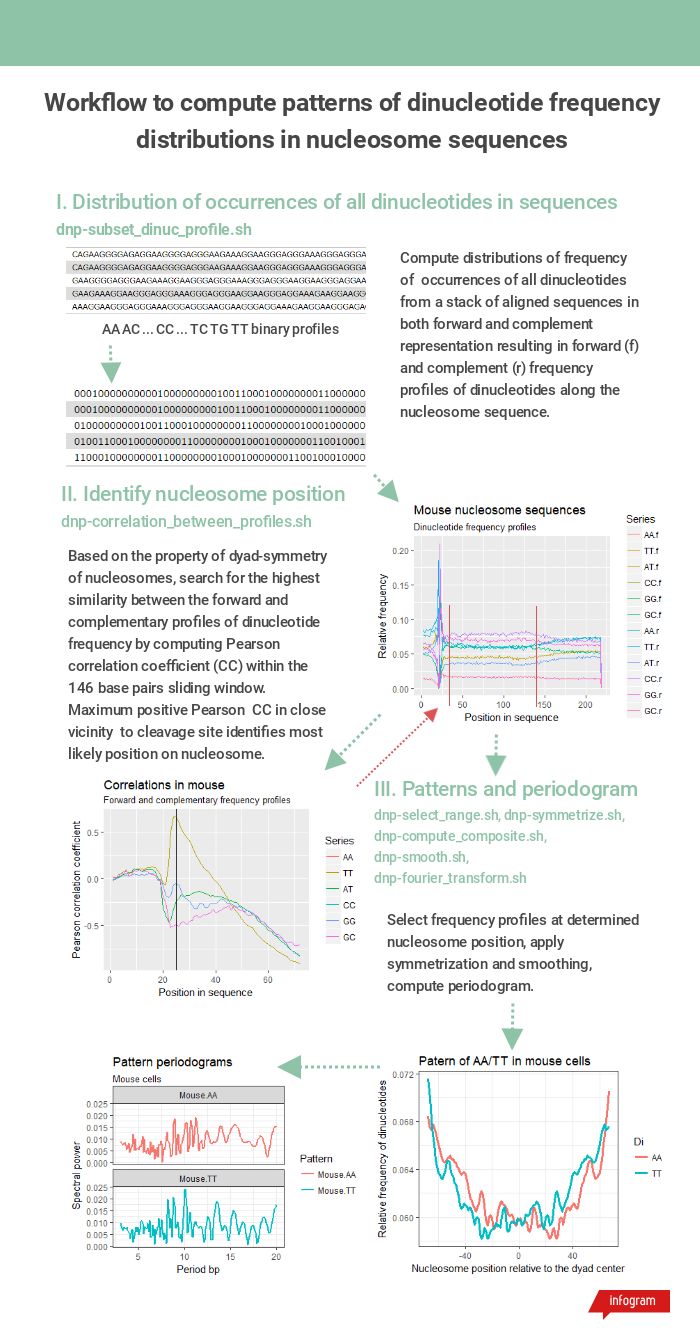
Figure 1. The workflow of dinucleotide frequency pattern computation from a batch of nucleosomes fasta sequences.
The whole dnpatterntools directory structure is following:
dnpatterntools/
├── bin
├── source
├── test
├── tools
│ ├── extra
│ └── test-data
└── tools-extra
├── bioconda-recipes
│ ├── dnp-binstrings
│ ├── dnp-corrprofile
│ ├── dnp-diprofile
│ └── dnp-fourier
└── ggplot-scripts
└── R
The bin and source folders contain binaries (might or might not not work on your system) and C++ code of core programs. The core programs are summarized below:
| C++ source | Name of binary | Description |
|---|---|---|
| binstrings.cpp | dnp-binstrings | Converts fasta sequence into binary string of dinucleotide occurrences |
| diprofile.cpp | dnp-diprofile | Computes profiles of dinucleotide frequency of occurrence in a batch of aligned fasta sequences |
| corrprofile.cpp | dnp-corrprofile | Computes Pearson correlation between a forward and reversed complement dinucleotide frequency profiles |
| Fourier_Transform.cpp | dnp-fourier | Computes either smoothed and normalized dinucleotide frequency profile or its periodogram |
The tools folder contains tools that implement complete workflow to obtain and characterize dinucleotide patterns in a batch of fasta sequences. The tools are written in shell and depends on the core tools. Each tool has an associated galaxy xml wrapper with the same name. The Galaxy wrappers have been tested and served using Planemo (to be submitted to the Galaxy ToolShed). Below is a summary of the tools:
| Script name | Galaxy tool name | Description |
|---|---|---|
| dnp-subset-dinuc-profile.sh | Dinucleotide frequencies | Computes frequencies of occurrence of a subset of dinucleotides in a batch of fasta |
| dnp-correlation-between-profiles.sh | Correlations | Computes Pearson correlation between a forward and reversed complement dinucleotide frequency profiles |
| dnp-select-range.sh | Select interval | Selects rows from the dinucleotide frequency profiles matrix within a give range |
| dnp-symmetrize.sh | Symmetrize | Applies symmetrization operation on forward and complement dinucleotide profiles |
| dnp-compute-composite.sh | Composite profiles | Computes composite dinucleotide frequency profiles |
| dnp-smooth.sh | Smooth | Applies smoothing and normalization on a given dinucleotide frequency profile |
| dnp-fourier-transform.sh | Periodogram | Computes periodogram for a give dinucleotide profile |
The test folder contains shell scripts of test calls to the core programs and dnp tools.
The tools-extra folder contains bioconda-recipes for the core tools. The ggplot-scripts contains R functions to visualize the tools outputs.
Download the repository or use git clone. Follow building instructions in the source folder. If core programs are already installed, then descend into test directory to run tests. Run the test-dependencies.sh to test the core programs. Run test_tools.sh to test tools. The test data files are in tools/test-data folder. A standard use is described in a workflow. However, these tools may have a wider scope of application.
dnp-binstrings:
binstrings - Binary strings from fasta
======================================
SYNOPSIS
binstrings [OPTIONS] "fastaFile.fa"
DESCRIPTION
This program reads the fasta file and each sequence is transformed into
0011 form in which ones denote dinucleotides and zeros elsewhere. Binary
sequence is printed.
REQUIRED ARGUMENTS
FASTA_FILE STRING
OPTIONS
-h, --help
Display the help message.
--version-check BOOL
Turn this option off to disable version update notifications of the
application. One of 1, ON, TRUE, T, YES, 0, OFF, FALSE, F, and NO.
Default: 1.
-di, --dinucleotide STRING
Dinucleotide that is to identify in fasta sequences One of AA, AC,
AG, AT, CA, CC, CG, CT, GA, GC, GG, GT, TA, TC, TG, and TT. Default:
CC.
--version
Display version information.
EXAMPLES
binstrings -di CC path/to/fasta/file.fa
Compute binary strings matching CC in fasta sequences.
OUTPUT
100000000111000 CC chr9:42475963-42476182 CCAGGCAGACCCCATA 4
binary string, CC, fasta id, DNA sequence, occurrences
VERSION
Last update: September 2018
binstrings version: 1.0
SeqAn version: 2.4.0
dnp-corrprofile:
corrprofile - Correlations between Dinucleotide Profiles
========================================================
SYNOPSIS
corrprofile [OPTIONS] "dinucleotideProfilesFile"
DESCRIPTION
This program computes correlations between the profiles of dinucleotide
frequency on forward and reverse complement sequences within a sliding
window.
REQUIRED ARGUMENTS
PROFILE_FILE STRING
OPTIONS
-h, --help
Display the help message.
--version-check BOOL
Turn this option off to disable version update notifications of the
application. One of 1, ON, TRUE, T, YES, 0, OFF, FALSE, F, and NO.
Default: 1.
-w, --window INTEGER
Sliding window size, < than length. In range [10..146]. Default: 10.
-n, --length INTEGER
Dinucleotide profile sequence length. In range [25..600]. Default:
600.
-v, --verbose
Print parameters and variables.
--version
Display version information.
EXAMPLES
corrprofile -w 146 -n 400 path/to/profiles/file
Compute correlations at each position in 400bp long profile within
the sliding 146bp window
OUTPUT
Column of correlation coefficients
between forward and reverse profile at each position
VERSION
Last update: April 2017
corrprofile version: 1.0
SeqAn version: 2.4.0
dnp-diprofile:
diprofile - Dinucleotide Frequency Profile
==========================================
SYNOPSIS
diprofile [OPTIONS] "fastaFile.fa"
DESCRIPTION
This program computes a profile of a frequency of occurrence of the
dinucleotide in a batch of fasta sequences aligned by their start
position.
REQUIRED ARGUMENTS
FASTA_FILE STRING
OPTIONS
-h, --help
Display the help message.
--version-check BOOL
Turn this option off to disable version update notifications of the
application. One of 1, ON, TRUE, T, YES, 0, OFF, FALSE, F, and NO.
Default: 1.
-di, --dinucleotide STRING
Dinucleotide to compute a frequency profile in fasta file. One of
AA, AC, AG, AT, CA, CC, CG, CT, GA, GC, GG, GT, TA, TC, TG, and TT.
Default: AA.
-sl, --seqlength INTEGER
Sequence length in fasta file. In range [25..600]. Default: 600.
-c, --complement
Perform computation on COMPLEMENTARY sequences of the strings in
fasta file.
-v, --verbose
Print parameters and variables.
--version
Display version information.
EXAMPLES
diprofile -sl 146 -di CT path/to/fasta/file.fa
Compute CT profile in fasta sequences of 146bp long
diprofile -sl 146 -di CT -c path/to/fasta/file.fa
Compute CT profile in sequence complements of fasta sequences of
146bp long
OUTPUT
Column of relative frequencies of dinucleotide occurrences at each
position along fasta sequences of given length --seqlength
VERSION
Last update: April 2017
diprofile version: 1.0
SeqAn version: 2.4.0
dnp-fourier:
Fourier transform and smoothing of input sequence
input parameters:
------------------------------------------------
-f input sequence
-o output table
-l length of window of smoothing
-n type of normalisation:
0 base normalization
1 linear normalization
2 quadratic normalization
-t type of output table:
1 normalization
2 smoothing
3 Fourier transform
S.Hosid 2008 - 2018