Prototyped Segmentation
virtualenv -p python3 venv
source venv/bin/activate
pip install -r requirements.txt
python3 train.py configs/all.ymlThe following package should be installed
- pip install git+https://github.com/chriamue/pytorch-semseg
- pip install gluoncv,mxboard, tensorboardx
then protoseg can be installed
pip install git+https://github.com/chriamue/protosegImages should be copied to ./data folder. Images for training to train folder, the masks to train_masks folder. Images for validation to val folder, the masks to val_masks folder. Images for testing into test folder.
Every run is stored in a config file like: config
There should exist multiple backends.
- gluoncv
- pytorch-semseg
gluoncv:
backend: gluoncv_backend
backbone: resnet50ptsemseg_unet:
backend: ptsemseg_backend
backbone: unet
classes: 2
width: 572
height: 572
mask_width: 388
mask_height: 388
orig_width: 768
orig_height: 768
gray_img: Trueptsemseg_segnet:
backend: ptsemseg_backend
backbone: segnet
classes: 2
width: 512
height: 512There are several options for augmentation. You can find a full list of augmentators in the docs of imgaug. Each augmentator will be executed with probability 0.5 .
Shape augmentation will be executed on image and mask.
shape_augmentation:
- Affine:
rotate: -15
- Affine:
rotate: 30
- Affine:
scale: [0.8, 2.5]
- Fliplr:
p: 1.0Image augmentation will be executed only on the image.
img_augmentation:
- GaussianBlur:
sigma: [0, 0.5]
- AdditiveGaussianNoise:
scale: 50Filters are functions to preprocess images. The functions have to consume an image first and then some parameters and have to return the processed image. Parameters can be given as list or as dictionary.
filters:
- 'cv2.Canny': [100,200]
- 'protoseg.filters.morphological.erosion':
kernelw: 5
kernelh: 5
iterations: 1There are some metrices available to measure on validation data.
metrices:
- 'pixel_accuracy': 'protoseg.metrices.accuracy.pixel_accuracy'
- 'mean_accuracy': 'protoseg.metrices.accuracy.mean_accuracy'
- 'dice': 'protoseg.metrices.dice.dice'
- 'jaccard': 'protoseg.metrices.jaccard.jaccard'
- 'iou': 'protoseg.metrices.iou.iou'A report as PDF file of the results can be created.
Running
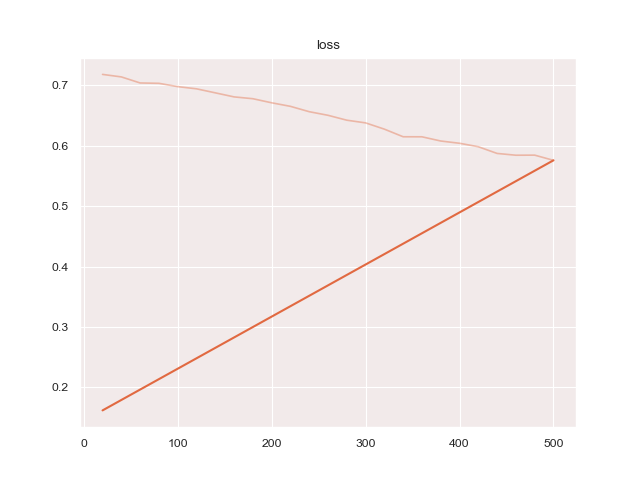
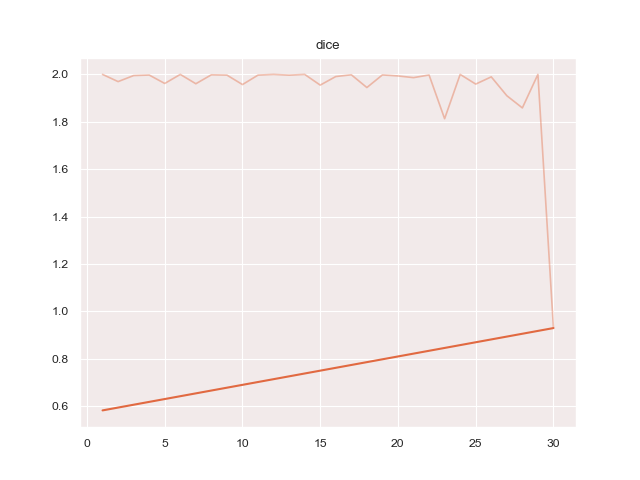
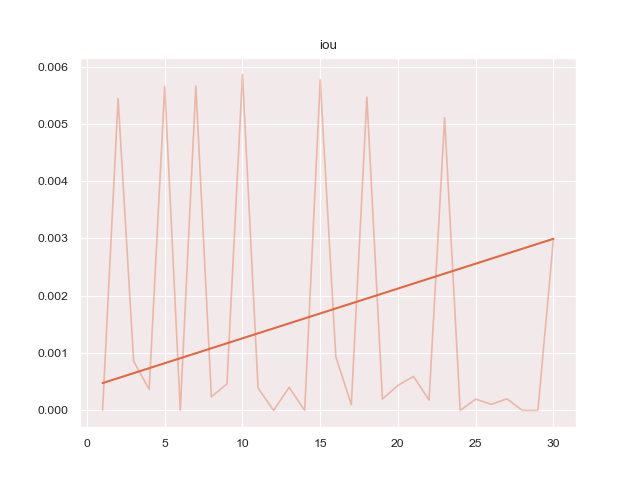
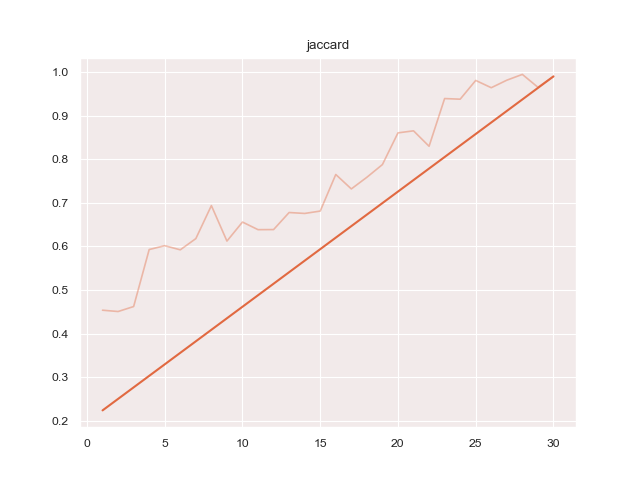
python3 train.py configs/ptsemseg_segnet.ymlproduces following images and pdf report file after training.
Hyperparameter trains multiple times with multiple configurations and tries to find
hyperparameters which produce the best loss.
Learnrate will be variated by default, but every other parameter in the config file can
be variated too.
An example configuration looks like:
ptsemseg_segnet:
flit: False
filters:
- 'cv2.Canny': [100,200]
hyperparamopt:
- flip: [True, False]
- filters: [['protoseg.filters.canny.addcanny': [100,200]],['protoseg.filters.morphological.opening': [5,5,1]]]A report will be generated as pdf file.
In folder ./scripts is ultrasound-nerve-segmentation.py which should be run as
python3 ./scripts/ultrasound-nerve-segmentation.py /path/to/competition-data data/The script extracts competition images and copies them to the data folder.
A 2017 Guide to Semantic Segmentation with Deep Learning Satellite Image Segmentation: a Workflow with U-Net