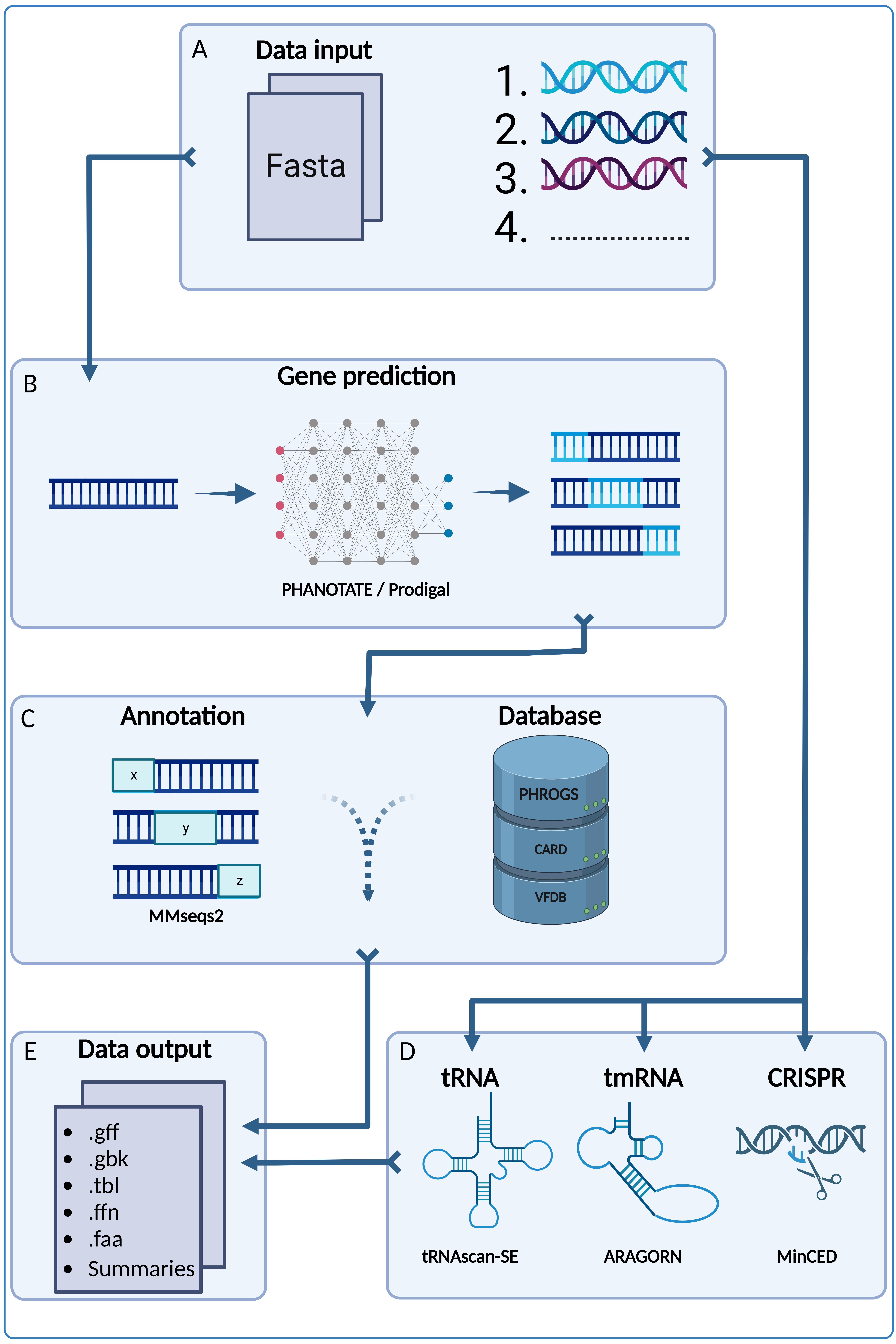
pharokka is a rapid standardised annotation tool for bacteriophage genomes and metagenomes.
If you are looking for rapid standardised annotation of bacterial genomes, please use prokka, which inspired the creation of pharokka, or bakta.
Check out the full documentation at https://pharokka.readthedocs.io.
pharokka has been recently published in Bioinformatics:
George Bouras, Roshan Nepal, Ghais Houtak, Alkis James Psaltis, Peter-John Wormald, Sarah Vreugde, Pharokka: a fast scalable bacteriophage annotation tool, Bioinformatics, Volume 39, Issue 1, January 2023, btac776, https://doi.org/10.1093/bioinformatics/btac776.
If you use pharokka, please see the full Citation section for a list of all programs pharokka uses, in order to fully recognise the creators of these tools for their work.
pharokka uses PHANOTATE, the only gene prediction program tailored to bacteriophages, as the default program for gene prediction. Prodigal is also available as an alternative. Following this, functional annotations are assigned by matching each predicted coding sequence (CDS) to the PHROGs, CARD and VFDB databases using MMseqs2. pharokka's main output is a GFF file suitable for using in downstream pangenomic pipelines like Roary. pharokka also generates a cds_functions.tsv file, which includes counts of CDSs, tRNAs, tmRNAs, CRISPRs and functions assigned to CDSs according to the PHROGs database. See the full usage and check out the full documentation for more details.
Pharokka v1.2.0 implements a major new feature. It quickly matches each input contig against the INPHARED database (paper) using mash distances, which may be useful if you are annotating novel phages or metagenomic input samples. If you use this feature, please make sure you cite INPHARED. Please see the Citation section for details.
v 1.2.0 also adds the ability to re-orient your phage specifying a coordinate and strandedness using the terminase large subunit reorientation mode, then annotate the re-oriented phage. Please see the usage section in the Documentation for more details.
To use v1.2.0, you will need to update your Pharokka database to include INPHARED by running install_databases.py -o <path/to/databse_dir>.
- pharokka
- Quick Start
- Installation
- Database Installation
- Beginner Conda Installation
- Usage
- Version Log
- System
- Time
- Benchmarking
- Bugs and Suggestions
- Citation
The easiest way to install pharokka is via conda:
conda install -c bioconda pharokka
Followed by database download and installation:
install_databases.py -o <path/to/databse_dir>
And finally annotation:
pharokka.py -i <phage fasta file> -o <output directory> -d <path/to/database_dir> -t <threads>
Please read below for more details, especially if you are an inexperienced command line user.
The easiest way to install pharokka is via conda. For inexperienced command line users, this method is highly recommended.
conda install -c bioconda pharokka
This will install all the dependencies along with pharokka. The dependencies are listed in environment.yml.
If conda is taking a long time to solve the environment, try using mamba:
conda install mamba
mamba install -c bioconda pharokka
Alternatively, the development version of pharokka (which may include new, untested features) can be installed manually via github.
git clone https://github.com/gbouras13/pharokka.git
The dependencies found in environment.yml will then need to be installed manually.
For example using conda to install the required dependencies:
git clone https://github.com/gbouras13/pharokka.git
cd pharokka
conda env create -f environment.yml
conda activate pharokka_env
And then to run pharokka (assuming you are still in the pharokka directory)
./bin/install_databases.py -h
./bin/pharokka.py -h
To install the pharokka database to the default directory:
install_databases.py -d
If you would like to specify a different database directory (recommended), that can be achieved as follows:
install_databases.py -o <path/to/databse_dir>
If this does not work, you an alternatively download the databases from Zenodo at https://zenodo.org/record/7563578/files/pharokka_v1.2.0_database.tar.gz and untar the directory in a location of your choice.
If you prefer to use the command line:
wget "https://zenodo.org/record/7563578/files/pharokka_v1.2.0_database.tar.gz"
tar -xzf pharokka_v1.2.0_database.tar.gz
which will create a directory called "pharokka_v1.2.0_database" containing the databases.
If you are new to using the command-line, please install conda using the following instructions.
- Install Anaconda. I would recommend miniconda.
- Assuming you are using a Linux x86_64 machine (for other architectures, please replace the URL with the appropriate one on the miniconda website).
curl -O https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
For Mac (Intel, will also work with M1):
curl -O https://repo.anaconda.com/miniconda/Miniconda3-latest-MacOSX-x86_64.sh
- Install miniconda and follow the prompts.
sh Miniconda3-latest-Linux-x86_64.sh
- After installation is complete, you should add the following channels to your conda configuration:
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
- After this, conda should be installed (you may need to restart your terminal). It is recommended that mamba is also installed, as it will solve the enviroment quicker than conda:
conda install mamba
- Finally, I would recommend installing pharokka into a fresh environment. For example to create an environment called pharokkaENV with pharokka installed:
mamba create -n pharokkaENV pharokka
conda activate pharokkaENV
install_databases.py -h
pharokka.py -h
Once the databases have finished downloading, to run pharokka:
pharokka.py -i <fasta file> -o <output directory> -t <threads>
To specify a different database directory (recommended):
pharokka.py -i <fasta file> -o <output directory> -d <path/to/database_dir> -t <threads> -p <prefix>
For a full explanation of all arguments, please see usage.
pharokka defaults to 1 thread.
usage: pharokka.py [-h] [-i INFILE] [-o OUTDIR] [-d DATABASE] [-t THREADS] [-f] [-p PREFIX] [-l LOCUSTAG] [-g GENE_PREDICTOR] [-m]
[-c CODING_TABLE] [-e EVALUE] [--terminase] [--terminase_strand TERMINASE_STRAND] [--terminase_start TERMINASE_START]
[-V] [--citation]
pharokka: fast phage annotation program
options:
-h, --help show this help message and exit
-i INFILE, --infile INFILE
Input genome file in fasta format.
-o OUTDIR, --outdir OUTDIR
Directory to write the output to.
-d DATABASE, --database DATABASE
Database directory. If the databases have been installed in the default directory, this is not required. Otherwise specify the path.
-t THREADS, --threads THREADS
Number of threads for mmseqs and hhsuite. Defaults to 1.
-f, --force Overwrites the output directory.
-p PREFIX, --prefix PREFIX
Prefix for output files. This is not required.
-l LOCUSTAG, --locustag LOCUSTAG
User specified locus tag for the gff/gbk files. This is not required. A random locus tag will be generated instead.
-g GENE_PREDICTOR, --gene_predictor GENE_PREDICTOR
User specified gene predictor. Use "-g phanotate" or "-g prodigal". Defaults to phanotate (not required unless prodigal is desired).
-m, --meta meta mode for metavirome input samples
-c CODING_TABLE, --coding_table CODING_TABLE
translation table for prodigal. Defaults to 11. Experimental only.
-e EVALUE, --evalue EVALUE
E-value threshold for mmseqs2 PHROGs database search. Defaults to 1E-05.
--terminase Runs "terminase large subunit" re-orientation mode. Single genome input only and requires --terminase_strand and --terminase_start to be specified.
--terminase_strand TERMINASE_STRAND
Strand of terminase large subunit. Must be "pos" or "neg".
--terminase_start TERMINASE_START
Start coordinate of the terminase large subunit.
-V, --version Print pharokka Version
--citation Print pharokka Citation
A brief description of what is new in each update of pharokka can be found in the HISTORY.md file.
pharokka has been tested on Linux and MacOS (M1 and Intel).
On a standard 16GB RAM laptop specifying 8 threads, pharokka should take between 3-10 minutes to run for a single phage, depending on the genome size.
pharokka (v1.1.0) has been benchmarked on an Intel Xeon CPU E5-4610 v2 @ 2.30 specifying 16 threads. Below is benchamarking comparing Pharokka run with PHANOTATE and Prodigal against Prokka v1.14.6 run with PHROGs HMM profiles, as modified by Andrew Millard (https://millardlab.org/2021/11/21/phage-annotation-with-phrogs/).
Benchmarking was conducted on Enterbacteria Phage Lambda (Genbank accession J02459) Staphylococcus Phage SAOMS1 (Genbank Accession MW460250) and 673 crAss-like phage genomes in one multiFASTA input taken from Yutin, N., Benler, S., Shmakov, S.A. et al. Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nat Commun 12, 1044 (2021) https://doi.org/10.1038/s41467-021-21350-w.
For the crAss-like phage genomes, Pharokka meta mode -m was enabled.
| Phage Lambda | pharokka PHANOTATE | pharokka Prodigal | Prokka with PHROGs |
|---|---|---|---|
| Time (min) | 4.19 | 3.88 | 0.27 |
| CDS | 88 | 61 | 62 |
| Coding Density (%) | 94.55 | 83.69 | 84.96 |
| Annotated Function CDS | 43 | 37 | 45 |
| Unknown Function CDS | 45 | 24 | 17 |
| Phage SAOMS1 | pharokka PHANOTATE | pharokka Prodigal | Prokka with PHROGs |
|---|---|---|---|
| Time (min) | 4.26 | 3.89 | 0.93 |
| CDS | 246 | 212 | 212 |
| Coding Density (%) | 92.27 | 89.69 | 89.31 |
| Annotated Function CDS | 92 | 93 | 92 |
| Unknown Function CDS | 154 | 119 | 120 |
| 673 crAss-like genomes from Yutin et al., 2021 | pharokka PHANOTATE Meta Mode | pharokka Prodigal Meta Mode | Prokka with PHROGs |
|---|---|---|---|
| Time (min) | 106.55 | 11.88 | 252.33 |
| Time Gene Prediction (min) | 96.21 | 3.4 | 5.12 |
| Time tRNA Prediction (min) | 1.25 | 1.08 | 0.3 |
| Time Database Searches (min) | 6.75 | 5.58 | 238.77 |
| CDS | 138628 | 90497 | 89802 |
| Contig Min Coding Density (%) | 66.01 | 46.18 | 46.13 |
| Contig Max Coding Density (%) | 98.86 | 97.85 | 97.07 |
| Annotated Function CDS | 9341 | 9228 | 14461 |
| Unknown Function CDS | 129287 | 81269 | 75341 |
pharokka scales well for large metavirome datasets due to the speed of mmseqs2. In fact, as the size of the input file increases, the extra time taken is required for running gene prediction (particularly PHANOTATE) and tRNA-scan SE2 - the time taken to conduct mmseqs2 searches remain small due to its many vs many approach.
If you require fast annotations of extremely large datasets (i.e. thousands of input contigs), running pharokka with Prodigal is recommended.
If you come across bugs with pharokka, or would like to make any suggestions to improve the program, please open an issue or email george.bouras@adelaide.edu.au
George Bouras, Roshan Nepal, Ghais Houtak, Alkis James Psaltis, Peter-John Wormald, Sarah Vreugde, Pharokka: a fast scalable bacteriophage annotation tool, Bioinformatics, Volume 39, Issue 1, January 2023, btac776, https://doi.org/10.1093/bioinformatics/btac776
If you use pharokka, I would recommend a citation in your manuscript along the lines of:
- All phages were annotated with Pharokka v ___ (Bouras, et al. 2023). Specifically, coding sequences (CDS) were predicted with PHANOTATE (McNair, et al. 2019), tRNAs were predicted with tRNAscan-SE 2.0 (Chan, et al. 2021), tmRNAs were predicted with Aragorn (Laslett, et al. 2004) and CRISPRs were preducted with CRT (Bland, et al. 2007). Functional annotation was generated by matching each CDS to the PHROGs (Terzian, et al. 2021), VFDB (Chen, et al. 2005) and CARD (Alcock, et al. 2020) databases using MMseqs2 (Steinegger, et al. 2017). Contigs were matched to their closest hit in the INPHARED database (Cook, et al. 2021) using mash (Ondov, et al. 2016).
With the following full citations for the constituent tools below:
- Cook R, Brown N, Redgwell T, Rihtman B, Barnes M, Clokie M, Stekel DJ, Hobman JL, Jones MA, Millard A. INfrastructure for a PHAge REference Database: Identification of Large-Scale Biases in the Current Collection of Cultured Phage Genomes. PHAGE. 2021. Available from: http://doi.org/10.1089/phage.2021.0007.
- McNair K., Zhou C., Dinsdale E.A., Souza B., Edwards R.A. (2019) "PHANOTATE: a novel approach to gene identification in phage genomes", Bioinformatics, https://doi.org/10.1093/bioinformatics/btz26.
- Chan, P.P., Lin, B.Y., Mak, A.J. and Lowe, T.M. (2021) "tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes", Nucleic Acids Res., https://doi.org/10.1093/nar/gkab688.
- Steinegger M. and Soeding J. (2017), "MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets", Nature Biotechnology https://doi.org/10.1038/nbt.3988.
- Ondov, B.D., Treangen, T.J., Melsted, P. et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17, 132 (2016). https://doi.org/10.1186/s13059-016-0997-x.
- Terzian P., Olo Ndela E., Galiez C., Lossouarn J., Pérez Bucio R.E., Mom R., Toussaint A., Petit M.A., Enault F., "PHROG : families of prokaryotic virus proteins clustered using remote homology", NAR Genomics and Bioinformatics, (2021), https://doi.org/10.1093/nargab/lqab067.
- Bland C., Ramsey L., Sabree F., Lowe M., Brown K., Kyrpides N.C., Hugenholtz P. , "CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats", BMC Bioinformatics, (2007), https://doi.org/10.1186/1471-2105-8-209.
- Laslett D., Canback B., "ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences.", Nucleic Acids Research (2004) https://doi.org/10.1093/nar/gkh152.
- Chen L., Yang J., Yao Z., Sun L., Shen Y., Jin Q., "VFDB: a reference database for bacterial virulence factors", Nucleic Acids Research (2005) https://doi.org/10.1093/nar/gki008.
- Alcock et al, "CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database." Nucleic Acids Research (2020) https://doi.org/10.1093/nar/gkz935.
- Larralde, M., (2022). Pyrodigal: Python bindings and interface to Prodigal, an efficient method for gene prediction in prokaryotes. Journal of Open Source Software, 7(72), 4296. doi:10.21105/joss.04296.