BranchedGP is a package for building Branching Gaussian process models in python, using TensorFlow and GPFlow. The model is described in the paper
This is now published in Genome Biology.
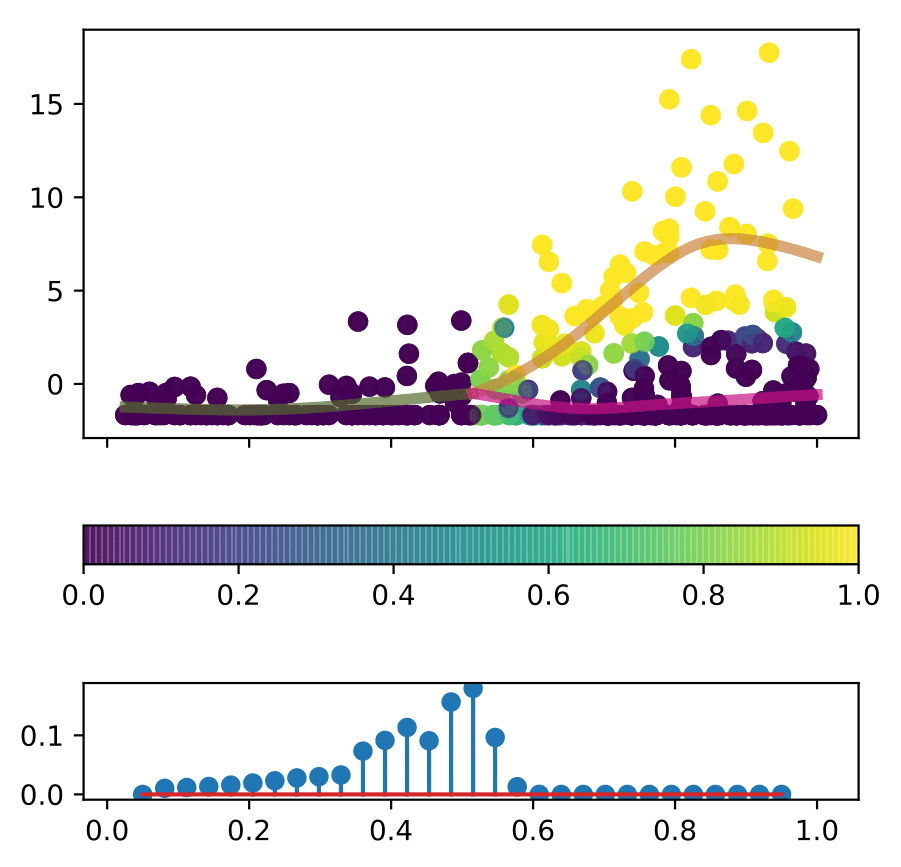
An example of what the model can provide is shown below.
- The posterior cell assignment is shown in top subpanel: each cell is assigned a probability of belonging to a branch.
- In the bottom subpanel the posterior branching time is shown: the probability of branching at a particular pseudotime.
This project requires Python3.7 or earlier (TensorFlow 1 requirement).
Create a virtual environment, activate it and run make install.
For a quick introduction see the notebooks/Hematopoiesis.ipynb notebook.
Therein we demonstrate how to fit the model and compute
the log Bayes factor for two genes.
The Bayes factor in particular is calculated by calling CalculateBranchingEvidence
after fitting the model using FitModel.
This notebook should take a total of 6 minutes to run.
| File name |
Description |
|---|---|
| Hematopoiesis | Application of BGP to hematopoiesis data. |
| SyntheticData | Application of BGP to synthetic data. |
| SamplingFromTheModel | Sampling from the BGP model. |
In the paper we compare the BGP model to the BEAM method proposed
in monocle 2. In monocle/runMonocle.R the R script for performing
Monocle and BEAM on the hematopoiesis data is included.
| File name |
Description |
|---|---|
| FitBranchingModel.py | Main file for user to call BGP fit, see function FitModel |
| pZ_construction_singleBP.py | Construct prior on assignments; use by variational code. |
| assigngp_dense.py | Variational inference code to infer function labels. |
| assigngp_denseSparse.py | Sparse inducing point variational inference code to infer function labels. |
| branch_kernParamGPflow.py | Branching kernels. Includes independent kernel as used in the overlapping mixture of GPs and a hardcoded branch kernel for testing. |
| BranchingTree.py | Code to generate branching tree. |
| VBHelperFunctions.py | Plotting code. |
- Tests:
make test - Install dependencies (into an active virtual environment):
make install - Format code:
make format - Run a jupyter notebook server:
make jupyter_server