This repository contains the code used in the experiments of Rommel, Paillard, Moreau and Gramfort, "Data augmentation for learning predictive models on EEG: a systematic comparison", 2022.
Start by cloning the repository:
git clone https://github.com/eeg_augmentation_benchmark/eeg_augmentation_benchmark-2022.gitExperiments within this repository use the data augmentations implemented in Braindecode library, which is based on the simple API from skorch wrapping up the pytorch deep learning framework. The repository also heavily relies on EEG preprocessing tools and datasets available in the MNE Python and MOABB libraries.
Other depencies are:
- joblib
- numpy
- pandas
- matplotlib
- seaborn
All of them are listed in environment.yml file and can be installed into a new environment using Anaconda by running:
conda env create -f environment.ymlfrom the package root directory.
After installing the necessary requirements, you should install the package itself. For this, go to the root directory of the package and run
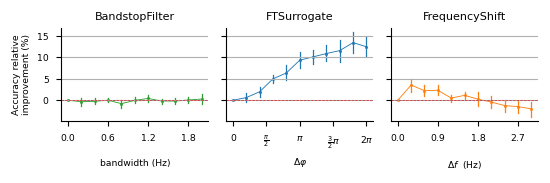
pip install -e .Parameter searches presented on figures 4, 9, 13 and 14 can be run with:
bash param_search/run_sleep.sh $DEVICE
bash param_search/run_bci.sh $DEVICEwhere $DEVICE corresponds to cpu or any cuda device that should be used for training.
Once these scripts are done executing, you can plot the corresponding figures with
bash param_search/plot_all.shThey will be saved in outputs/physionet/figures/ and outputs/BCI/figures/.
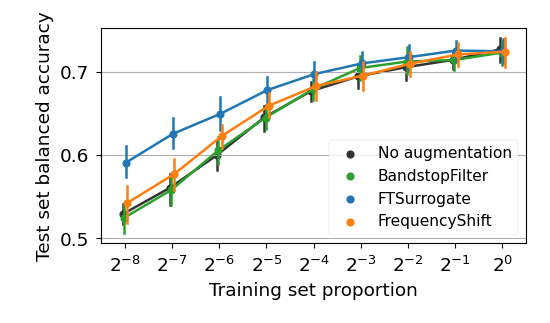
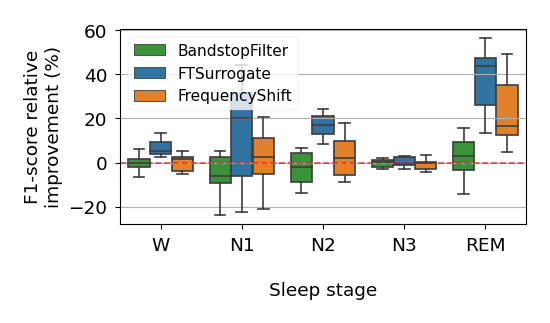
Likewise, to plot the learning curves from figures 5, 10, 15, 16 and 19, as well as the boxplots 6, 11, 12, 17 and 18, one can run:
bash learning_curves/run_sleep.sh $DEVICE
bash learning_curves/run_bci.sh $DEVICE
bash learning_curves/plot_all.shIf you use the code in this repository, please consider citing:
@article{rommel-et-al-2022,
title={Data augmentation for learning predictive models on {EEG}: a systematic comparison},
author={Rommel, Cédric and Paillard, Joseph and Moreau, Thomas and Gramfort, Alexandre},
year={2022},
journal={arXiv},
}