ScaffoldGVAE: A Variational Autoencoder Based on Multi-View Graph Neural Networks for Scaffold Generation and Scaffold Hopping of Drug Molecules
You can use the environment.yml file to create a new conda environment with all the necessary dependencies for ScaffoldGVAE.
git clone git@github.com:ecust-hc/ScaffoldGVAE.git
cd ScaffoldGVAE
conda env create -f environment.yml
conda activate ScaffoldGVAE
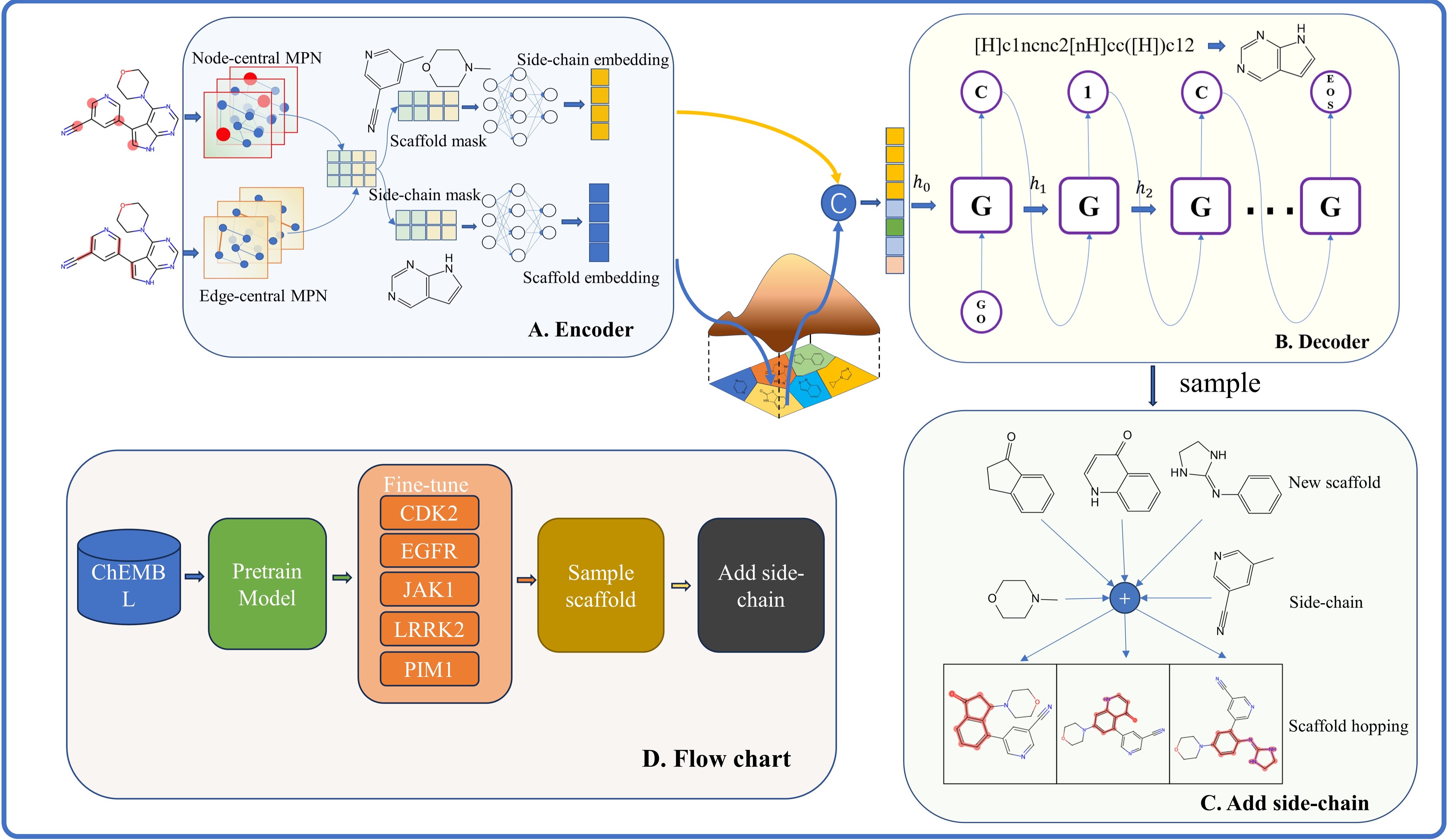
ScaffoldGVAE includes three sub-modules:
-
Sca_extraction.py: The molecular scaffolds was extracted and the data set was constructed. -
Train.py: Used for pre-training on big dataset. -
fine_tuning.py: Used for fine-tuning the pre-trained neural network on the known bioactive compounds against specific protein targets. -
sample.py: Given a reference molecule and its corresponding scaffold ,sampling new scaffolds and adding side-chain for scaffold hopping.
Example of running the command:
python Sca_extraction.py
python pre_train.py
python fine_tuning.py
python sample.py