FragHub is a powerful tool designed to standardize and organize mass spectrometry (MS) data from OMSLs (Open Mass Spectra Libraries). The main objective of FragHub is to simplify and improve the process of MS data analysis by providing standardized, consistent, and easily accessible data.
Key features:
- Data standardization: FragHub standardizes field names and values of MS spectra from various databases, ensuring data consistency and compatibility.
- Optional peak list filtering: FragHub applies filters to peak lists to streamline them by retaining only essential data, significantly reducing data size without compromising relevance for analysis.
- Recalculation and normalization of chemical identifiers: The program recalculates and normalizes chemical identifiers such as SMILES, InChI, and InChIKey, ensuring a uniform representation of molecular structures.
- Spectra organization: FragHub separates spectra based on different experimental parameters such as polarity (positive/negative), chromatographic mode (LC/GC), and acquisition type (experimental/in silico), facilitating their use and subsequent analysis.
- Compatibility with various analysis software: Standardized spectra produced by FragHub are compatible with multiple analysis software, including MSdial, MZmine, and Flash Entropy Search, providing users with maximum flexibility in choosing analysis tools.
Warning: All spectra deemed inconsistent, i.e., those lacking SMILES and InChI, precursor m/z, and adduct information, are removed during the processing by FragHub.
To install all dependencies, double-click on the install script corresponding to your OS.
NB: Make sure that python is in the path variables and that you run Python >= 3.9
To use this programme:
- Double-click on your corresponding OS run script into scripts folder.
NB: If you have a file that contains only In-Silico spectra AND this is not specified within the filename or the spectrum, you can simply suffix the filename with "_insilico", like this: "UNPD_ISDB_R_p01_insilico.mgf".
>NB: If you have a file that contains **only** GC-IE spectra AND this is not specified within the filename or the spectrum, you can simply suffix the filename with "_GC_IE", like this: "GMD_20111121_MDN35_ALK_MSP_GC_IE.msp".
- FragHub GUI start
- First tab: Select the folder where your spectral data files are stored ('.msp', '.json', '.csv', '.mgf').
- Second tab: Select the folder in which you want your data to be saved (FragHub automatically creat a sub-folder named like the project selected/created)
- Third tab: This area allows users to select specific functions for inclusion during the processing stage. Moreover, it provides the option to adjust the respective parameters of each function.
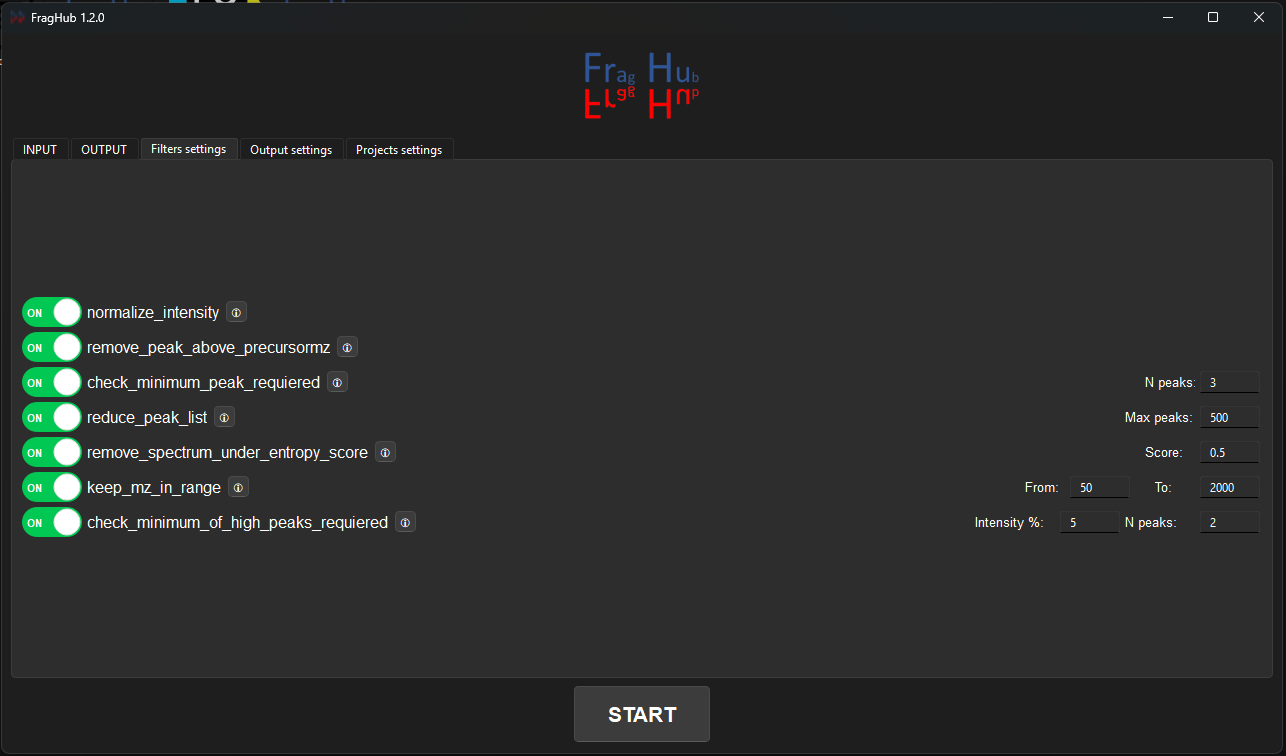
- Fourth tab: Select the output file format of your preference. By default, all formats are selected.
- Fifth tab: This tab facilitates the management of distinct projects (like 'internal lab standards' or 'In-Silico DB', etc). Either select from a previously created project or create a new one by just entering your desired project name.

- Sixth tab: The 'Reset updates' option, when checked, allows for a reset of everything related to previously encountered spectra from the current selected profil. This will also delete all existing files located in the OUTPUT/{current selected profil} folder.

- First tab: Select the folder where your spectral data files are stored ('.msp', '.json', '.csv', '.mgf').
- When the execution is complete, please remember to take a copy of your cleaned files from the OUTPUT folder and place them in a different location.
- DO NOT DELETE FILES INTO 'OUTPUT' AFTER COPY CLEANED VERSIONS.
check_minimum_peak_requiered(peak_array, n_peaks)
This function checks whether a given mass spectrum contains a minimum number of peaks. If the spectrum contains fewer peaks than the minimum requirement, it ignores the spectrum.
remove_peak_above_precursormz(peak_array, precursormz)
This function removes all peaks from the spectrum whose m/z value is greater than the precursor's m/z value plus 5 Da.
reduce_peak_list(peak_array, max_peaks)
This function reduces the peak list to a specified maximum number of peaks. The peaks to retain are chosen based on their intensity, with peaks of greater intensity being selected.
normalize_intensity(peak_array)
This function normalizes the intensity of all the peaks in a given spectrum to the maximum intensity.
keep_mz_in_range(peak_array, mz_from, mz_to)
This function takes an array of peak data (representing mass-to-charge ratio, or m/z) and returns a new array containing only those peaks whose m/z value falls between mz_from and mz_to.
check_minimum_of_high_peaks_requiered(peak_array, intensity_percent, no_peaks)
This function is used to check whether a given array containing peak data has a required minimum number of "high peaks". A "high peak" is defined as a peak whose intensity is above a certain percentage (intensity_percent) of the maximum intensity. If the array does not contain a sufficient number of "high peaks", the function ignore the spectrum.
- CSV files need to be separated by ';' with quotechar '"'.
- peaks columns need to be named 'peaks'.
- 'peaks' column need to be formatted with one of the following format, in string:
-
"[[79.054840, 12486.074219], [79.629868, 854.089905]]"
-
"
57.07042529 0.7697591662
71.08607535 1.507457981
97.06533991 0.4893302623
99.08098997 0.4737337839
137.09664 0.498920401
165.0915547 0.4243093978
"
-
If you wish to visualize the spectra tracing eliminated during the process, you can, from the TOOLS branch, start FragHub in the TOOLS\spectrum_loss_TRACES folder, and visualize the tracing from the jupyter notbook TOOLS\spectrum_loss_TRACES\TRACKERS.ipynb


