Matlab and shell scripts associated with the paper "Correcting datasets leads to more homogeneous early 20th century sea surface warming" by Duo Chan, Elizabeth C. Kent, David I. Berry, and Peter Huybers.
Most of these codes are Matlab .m files . We provide a script here for fast reproduction of Figures and Table in the main text. If you are reproducing the full analysis, which takes more computational resources and time to run, we provide two shell scripts as templates that runs the appropriate Matlab codes on clusters. The provided shell template is for submitting jobs on the Harvard Odyssey Cluster that uses a SLURM workload manager.
If you have issues implementing the above scripts, or identify any deficiencies, please contact Duo Chan (duochan@g.harvard.edu).
- Get started
- Quick reproduction of Figures and Tables
- Overview and system requirements
- A. Preprocess
- B. Main Code
- C. Figures and Tables
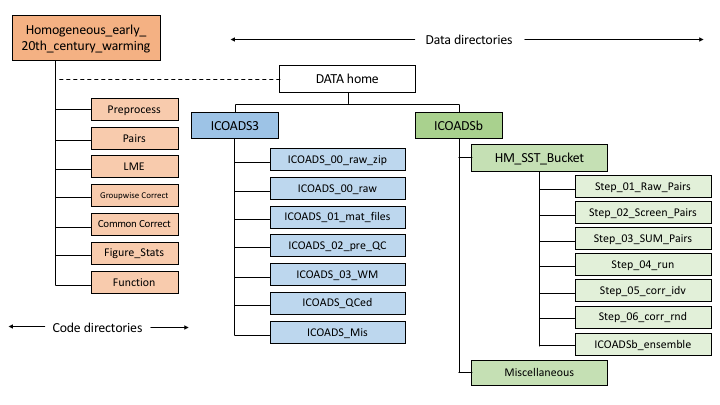
Run Chan_et_al_2019_init.m to initialize the analysis. This script will add all codes in this package to the Matlab path and set up directories structured following the below figure. The default path will be the directory of this package; so make sure that enough disk space is available (~1GB for simply reproducing figures and tables and ~350GB for full reproduction), or, specify another directory to store the data:
Chan_et_al_2019_init($DATA_home)
After initialization, run Quick_reproduction.m, a wrapper that downloads key results and then generates Figs. 1-4 and Table 1, as well as the numbers reported in the main text. Size of downloaded data is about 1GB and reproduction takes about 2 minutes to run on a laptop.
Quick_reproduction
Data downloaded are listed below:
-
SUM_corr_idv_HM_SST_Bucket_GC_*.mat: key statistics for ICOADSa and ICOADSb, which should be placed in
$DATA_home/ICOADSb/HM_SST_Bucket/. Key statistics include the spatial pattern of 1908-1941 trends, monthly SSTs over the North Pacific and North Atlantic, monthly SSTs near East Asia and the Eastern U.S., and PDO indices. -
SUM_corr_rnd_HM_SST_Bucket_GC_*.mat: as above but for 1,000 ICOADSb correction members.
-
1908_1941_Trd_TS_and_pdo_from_all_existing_datasets_20190424.mat: key statistics for existing major SST estimates, which should be placed in
$DATA_home/ICOADSb/Miscellaneous/. Major SST estimates are ERSST5, COBESST2, HadISST2, and HadSST3. All of these datasets are regridded to 5-degree resolution, which can be downloaded as a part of the supporting data. -
Stats_HM_SST_Bucket_deck_level_1.mat: statistics of numbers of measurements for each nation-deck group, which should be placed in
$DATA_home/ICOADSb/HM_SST_Bucket/. -
LME_HM_SST_Bucket_*.mat: groupwise offset estimates for bucket SSTs, which should be placed in
$DATA_home/ICOADSb/HM_SST_Bucket/Step_04_run/. -
CRUTEM.4.6.0.0.anomalies.nc: CRUTEM4 dataset, which should be placed in
$DATA_home/ICOADSb/Miscellaneous/.
Dependency is the Matlab m_map toolbox and we put a copy in this package.
________________________________
For users interested in reproducing our full analysis, we provide the following guidance to run the codes.
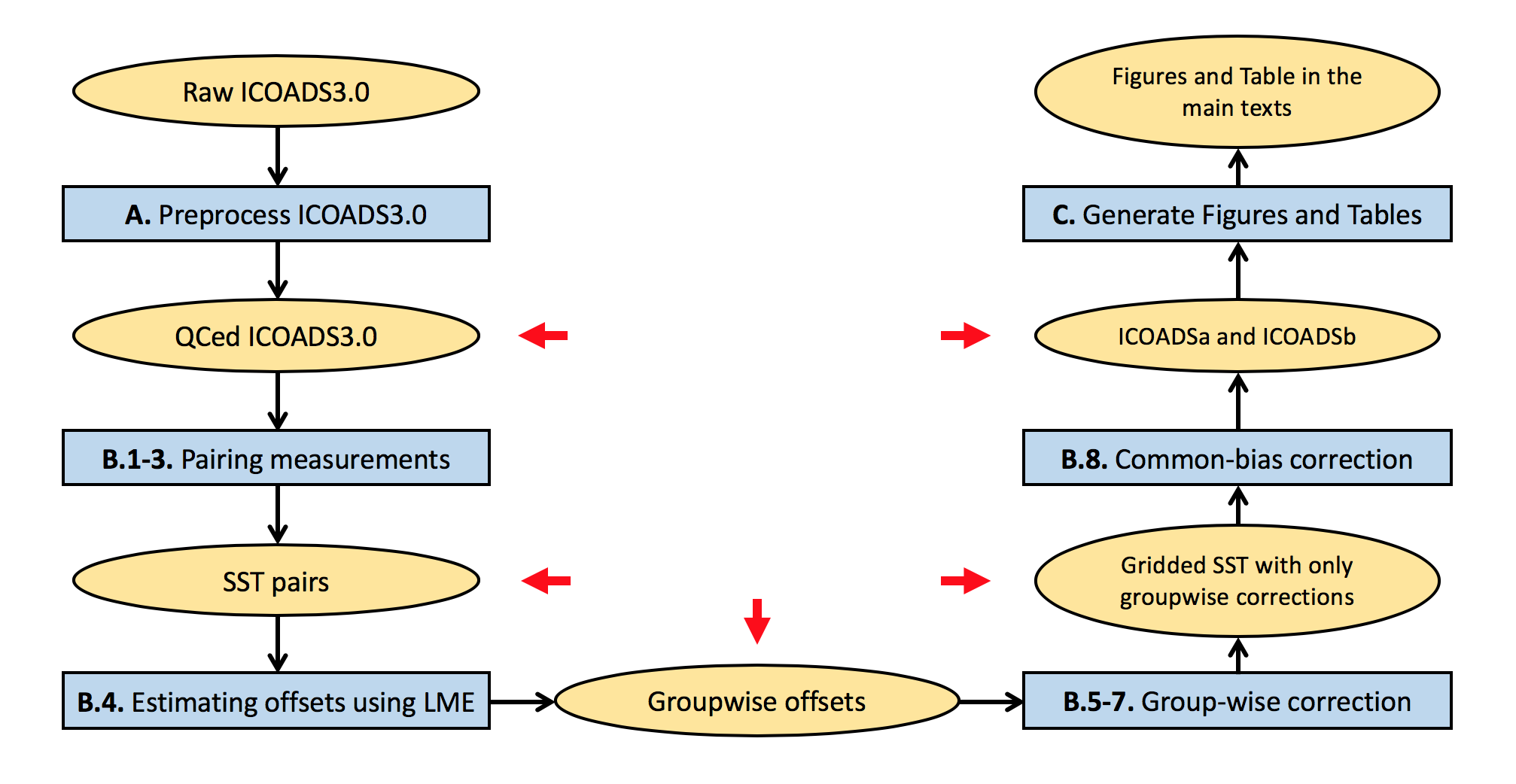
Below is the flow chart of the full analysis.
Several processing steps are memory and computationally intensive. Our analysis was run on Harvard Research Computing clusters and used 1200 CPUs and one analysis uses 150GB memory per CPU, and required 6,000 core-hours of computation and 350GB of disk space.
To reproduce our analysis, run get_supporting_data.m, a script that downloads supporting data files, unzips and moves these files to their target directories.
get_supporting_data
For purposes of facilitating reproduction we have also provided files resulting from our computation at various stages of the analysis (indicated by red arrows and see below for more details). One can choose to start from any of these checkpoints. Run get_check_points.m, a script that downloads checkpoint files, unzips and moves these files to their target directories.
get_check_points
This folder contains scripts for downloading and preprocessing the ICOADS3.0 data.
Ensure that 270GB of disk space is available for raw ICOADS3.0 and outputs during preprocessing steps, and we recommend to use a cluster. Or you can skip preprocessing by downloading the preprocessed .mat files (35GB) and place them in $DATA_home/ICOADS3/ICOADS_QCed/.
[Prerequisite] Ensure the following data and metadata are downloaded:
- ICOADS3.0 raw data: zipped ICOADS3.0 (28GB) are from RDA dataset 548.0. Please download files to
$DATA_home/ICOADS3/ICOADS_00_raw_zip/. We have also archived raw data we used here. After downloading, unzip files and place them in$DATA_home/ICOADS3/ICOADS_00_raw/. Note that unzipped ICOADS3.0 files are 153GB in total.
The following files belong to the supporting data and should be placed in $DATA_home/ICOADS3/ICOADS_Mis/.
-
OI_clim_1982_2014.mat: 33-year OI-SST daily climatology,
-
ERA_interim_AT2m_1985_2014_daily_climatology.mat: 30-year ERA-interim 2m-air temperature daily climatology,
-
Dif_DMAT_NMAT_1929_1941.mat: mean daytime and nighttime marine air temperature difference in 1929-1941,
-
Dif_DMAT_NMAT_1947_1956.mat: mean daytime and nighttime marine air temperature difference in 1947-1956,
-
Buddy_std_SST.mat: an output from step A.4,
-
Buddy_std_NMAT.mat: an output from step A.4.
Submit_preprocess.sh wraps all preprocessing steps and runs scripts on the Harvard Odyssey Cluster that uses a SLURM workload manager. If you are using a different machinery, please make necessary changes.
To perform the preprocessing, run
./Preprocess/Submit_preprocess.sh
Preprocessing contains five steps, as described below. Note that step A.2 follows Chan et al., submitted for SST and Kent et al. (2013) for nighttime marine air temperature, and steps A.3-A.5 follow Rayner et al. (2006) in performing buddy checking.
A.1. ICOADS_Step_01_ascii2mat.m converts ICOADS3.0 data from ASCII format to .mat files and stores them in $DATA_home/ICOADS3/ICOADS_01_mat_files/.
A.2. ICOADS_Step_02_pre_QC.m assigns missing country information and measurement method and outputs files to
$DATA_home/ICOADS3.O/ICOADS_02_pre_QC/.
A.3. ICOADS_Step_03_WM.m computes winsorized mean of 5-day SST at 1 degree resolution. These gridded data are stored in $DATA_home/ICOADS3/ICOADS_03_WM/.
A.4. ICOADS_Step_04_Neighbor_std.m computes between neighbor standard deviation of SST for each month.
A.5. ICOADS_Step_05_Buddy_check.m performs buddy check and other quality controls. Outputs are preprocessed files stored in $DATA_home/ICOADS3/ICOADS_QCed/.
As shown in the flow chart, this step contains pairing SST measurements, estimating offsets using LME, correcting groupwise offsets and gridding, and merging with common bucket corrections. These main steps can be accessed without preprocessing ICOADS3.0 by downloading the preprocessed .mat files(35GB) and place them in $DATA_home/ICOADS3/ICOADS_QCed/. When using command lines, we suggest:
wget -O $target_directory/ICOADS_QCed.tar.gz https://dataverse.harvard.edu/api/access/datafile/3424401
In case of use in deploying these scripts on a cluster, we provide Submit_main.sh, which runs the appropriate scripts on the Harvard Odyssey Cluster that uses a SLURM workload manager. To run the main analysis using the shell script, simply run (the command may vary on different machineries):
./Submit_main.sh
We strongly encourage you to go through the following documentation for prerequisites of individual steps and details of the workflow.
The Pairs folder contains functions that pair SST measurements.
[Prerequisite] Ensure that you have the following data or metadata placed in corresponding directories.
-
IMMA1_R3.0.0_YYYY-MM_QCed.mat: preprocessed ICOADS3.0 .mat files from running step A.1 - A.5. They should be placed in
$DATA_home/ICOADS3/ICOADS_QCed/and can be downloaded from here. -
DA_SST_Gridded_BUOY_sum_from_grid.mat and Diurnal_Shape_SST.mat: diurnal cycle estimates based on ICOADS3.0 buoy data. They belong to the supporting data and should be placed in
$DATA_home/ICOADSb/Miscellaneous/.
First, run HM_Step_01_02_Run_Pairs_dup.m to pair SST measurements following Chan and Huybers (2019). This script first calls HM_pair_01_Raw_Pairs_dup.m to pair SST measurements within 300km and 2days of one another and then calls HM_pair_02_Screen_Pairs_dup.m to screen pairs such that each measurement is used at most once. Output files are stored in $DATA_home/ICOADSb/HM_SST_Bucket/Step_01_Raw_Pairs/ and $DATA_home/ICOADSb/HM_SST_Bucket/Step_02_Screen_Pairs/.
Second, run HM_Step_03_SUM_Pairs_dup.m to combine screened pairs into one file, which will be used in following steps. The combined file, SUM_HM_SST_Bucket_Screen_Pairs_*.mat, is in $DATA_home/ICOADSb/HM_SST_Bucket/Step_03_SUM_Pairs/. It is a checkpoint.
The LME folder contains scripts that compute offsets among nation-deck groups of SST measurements using a linear-mixed-effect model (Chan and Huybers., 2019).
[Prerequisite] Ensure that you have the following data or metadata placed in corresponding directories.
-
SUM_HM_SST_Bucket_Screen_Pairs_*.mat: all paired measurements, which is the output of step B.3 and should be placed in
$DATA_home/ICOADSb/HM_SST_Bucket/Step_03_SUM_Pairs/. It is a checkpoint. -
OI_SST_inter_annual_decorrelation_20180316.mat: SST covariance structures estimated from the 33-year OI-SST dataset. It is one of the supporting data and should be placed in
$DATA_home/ICOADSb/Miscellaneous/.
Run HM_Step_04_LME_cor_err_dup.m to perform offset estimation. This script first calls HM_lme_bin_dup.m to aggregate SST pairs according to combinations of groupings, years, and regions, which reduces the number of SST differences from 17.8 million to 71,973. This step will output BINNED_HM_SST_Bucket_*.mat to directory $DATA_home/ICOADSb/HM_SST_Bucket/Step_04_run/.
HM_Step_04_LME_cor_err_dup.m then calls HM_lme_fit_hierarchy.m to fit the LME regression model and output groupwise offset estimates. The output file, LME_HM_SST_Bucket_*.mat, is a checkpoint and will also be placed in $DATA_home/ICOADSb/HM_SST_Bucket/Step_04_run/. Note that fitting the LME model involves inversion of a big matrix (~70,000 x 70,000) and takes 150GB of memory to run.
The Groupwise_Correct folder contains scripts that apply groupwise corrections and generates 5x5-degree gridded SST estimates. Groupwise corrections are applied to each SST measurement by removing offset estimated in step B.4 according to group, year, and region.
[Prerequisite] Ensure that you have the following data or metadata placed in corresponding directories.
-
IMMA1_R3.0.0_YYYY-MM_QCed.mat: preprocessed ICOADS3.0 .mat files from running step A.1 - A.5. They should be placed in
$DATA_home/ICOADS3/ICOADS_QCed/and can be downloaded from here. -
LME_HM_SST_Bucket_*.mat: groupwise offset estimates using an LME method, which is the output of step B.4 and should be placed in
$DATA_home/ICOADSb/HM_SST_Bucket/Step_04_run/. It is a checkpoint. -
internal_climate_patterns.mat: spatial pattern of PDO in SST. It is one of the supporting data and should be placed in
$DATA_home/ICOADSb/Miscellaneous/. -
nansum.m: a function that computes summation but returns NaN when all inputs are NaNs. The default Matlab nansum.m(release 2018b) function returns zero when all entries are NaNs. Our nansum.m is in
$home_Code/Function/.
First, run HM_Step_05_Corr_Idv.m to perform corrections using the maximum likelihood estimates of offsets and generate gridded SST estimates. HM_Step_05_Corr_Idv.m also generates gridded SST estimates that correct for only one group at a time. This step generates SST datasets that only have groupwise corrections, i.e., corr_idv_HM_SST_Bucket_*.mat in $DATA_home/ICOADSb/HM_SST_Bucket/Step_05_corr_idv/.
Then, run HM_Step_06_Corr_Rnd.m to generate a 1000-member ensemble of gridded SSTs, which can be used to estimate uncertainties of groupwise corrections. Corrections offsets are drawn from a multivariate normal distribution that centers on the maximum likelihood estimate (see appendix in Chan and Huybers., 2019). This step generates SST datasets that only have randomized groupwise corrections, i.e., corr_rnd_HM_SST_Bucket_*.mat in $DATA_home/ICOADSb/HM_SST_Bucket/Step_06_corr_rnd/.
Finally, run HM_Step_07_SUM_Corr.m to compute statistics of gridded SST estimates. These statistics include the spatial pattern of 1908-1941 trends, monthly SSTs over the North Pacific and North Atlantic, monthly SSTs near East Asia and the Eastern U.S., and PDO indices. This step will output SUM_corr_idv_HM_SST_Bucket_*.mat (central estimates) and SUM_corr_rnd_HM_SST_Bucket_*.mat (uncertainty estimates) in $DATA_home/ICOADSb/HM_SST_Bucket/. The two files are downloaded as checkpoints.
The Global folder contains scripts that merge large-scale common bucket corrections to raw ICOADS3.0 and ICOADS3.0 with groupwise corrections. The resulting datasets are called ICOADSa and ICOADSb, respectively.
[Prerequisite] Ensure that you have the following data or metadata placed in corresponding directories.
-
corr_idv_HM_SST_Bucket_*en_0*.mat: SST with only groupwise corrections using maximum likelihood estimates of offsets, which is an output from step B.5 and should be placed in
$DATA_home/ICOADSb/HM_SST_Bucket/Step_05_corr_idv/. It is a checkpoint. -
SUM_corr_rnd_HM_SST_Bucket_*.mat and SUM_corr_idv_HM_SST_Bucket_*.mat: key statistics of SST with only groupwise corrections, which are outputs from step B.6 and should be placed in
$DATA_home/ICOADSb/HM_SST_Bucket/. The two files are downloaded as checkpoints. -
Global_Bucket_Correction_start_ratio_35_mass_small_0.65_mass_large_1.7_start_ratio_35.mat: our reproduced 1850-2014 common bucket bias correction. The reproduction follows Kennedy et al., (2012) and details can be found in Extended Data Fig.8 and Supplemental Information Table 3 in Chan et al., submitted. These corrections involve running two bucket models, which can be accessed from here. Common bucket correction is a one of the supporting data and should be placed in
$DATA_home/ICOADSb/Miscellaneous/.
Following files are results of existing major SST estimates, i.e., ERSST5, COBESST2, HadISST2, and HadSST3. These files belong to the supporting data and should be placed in $DATA_home/ICOADSb/Miscellaneous/.
-
1908_1941_Trd_TS_and_pdo_from_all_existing_datasets_20190424.mat: key statistics for existing major SST estimates. We use HadSST3 ensemble members to estimate uncertainties associated with common bucket bias corrections.
-
All_earlier_SST_restimates_regridded_to_5x5_grids.mat. We use HadSST3 ensemble members to estimate uncertainties associated with common bucket bias corrections.
To perform step B.8, run HM_Step_08_Merge_GC.m. The script first calls GC_Step_01_SST_merge_GC_to_ICOADSb.m to generate ICOADSa and ICOADSb and then calls GC_Step_02_add_GC_to_ICOADSa_statistics.m to incorporate key statistics associated with common bucket corrections to SUM_corr_idv_HM_SST_Bucket_*.mat and SUM_corr_rnd_HM_SST_Bucket_*.mat.
This step will generate the following files in $DATA_home/ICOADSb/HM_SST_Bucket/, which will be used to generate Tables and Figures and can be downloaded from here.
-
ICOADS_a_b.mat: 5x5-degree gridded bucket SST datasets. ICOADSa contains only common bucket corrections, whereas ICOADSb contains both common bucket corrections and groupwise corrections.
-
SUM_corr_idv_HM_SST_Bucket_GC_*.mat: key statistics for ICOADSa and ICOADSb, see step B.7.
-
SUM_corr_rnd_HM_SST_Bucket_GC_*.mat: key statistics for 1,000 ICOADSb correction members.
This step will be the same as the quick reproduction that generates Figs. 1-4 and Table 1, as well as numbers reported in the main text.
Acknowledgement: We thank Packard Chan for his help in developing this page and checking scripts using his machinery.
Maintained by Duo Chan (duochan@g.harvard.edu)
Last Update: May 2, 2019