This package provides pre-trained U-net models for lung segmentation. For now, two models are available:
-
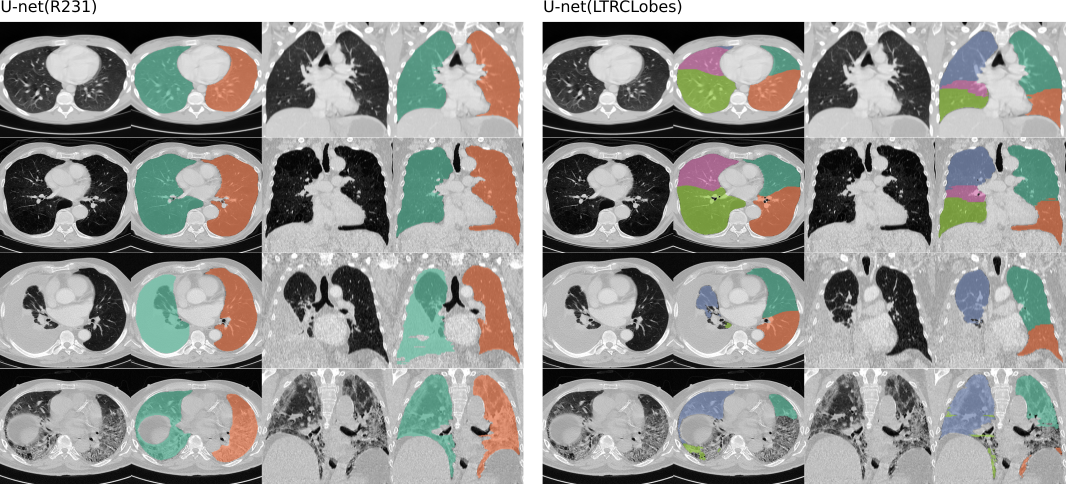
U-net(R231): This model was trained on a large and diverse dataset that covers a wide range of visual variabiliy. The model performs segmentation on individual slices, extracts right-left lung seperately includes airpockets, tumors and effusions. The trachea will not be included in the lung segmentation.
-
U-net(LTRCLobes): This model was trained on a subset of the LTRC dataset. The model performs segmentation of individual lung-lobes but yields limited performance when dense pathologies are present.
Examples of the two models applied. Left: U-net(R231), will distinguish between left and right lung and include very dense areas such as effusions (third row), tumor or sever fibrosis (fourth row) . Right: U-net(LTRLobes), will distinguish between lung lobes but will not include very dense areas.
For more exciting research on lung CT data, checkout the website of our research group: https://www.cir.meduniwien.ac.at/research/lung/
If you use this code or one of the trained models in your work please refer to:
Johannes Hofmanninger, Forian Prayer, Jeanny Pan, Sebastian Röhrich, Helmut Prosch and Georg Langs. "Automatic lung segmentation in routine imaging is a data diversity problem, not a methodology problem". 1 2020, https://arxiv.org/abs/2001.11767
This paper contains a detailed description of the dataset used, a thorough evaluation of the U-net(R231) model, and a comparison to reference methods.
pip install git+https://github.com/JoHof/lungmask
lungmask INPUT OUTPUT
If INPUT points to a file, the file will be processed. If INPUT points to a directory, the directory will be searched for DICOM series. The largest volume found (in terms of number of voxels) will be used to compute the lungmask. OUTPUT is the output filename. All ITK formats are supported.
Choose a model:
The U-net(R231) will be used as default. However, you can specify an alternative model such as LTRCLobes...
lungmask INPUT OUTPUT --modeltype unet --modelname LTRCLobes
For additional options type:
lungmask -h
from lungmask import lungmask
import SimpleITK as sitk
input_image = sitk.ReadImage(INPUT)
segmentation = lungmask.apply(input_image) # default model is U-net(R231)
input_image has to be a SimpleITK object.
Load an alternative model like so:
model = lungmask.get_model('unet','LTRCLobes')
segmentation = lungmask.apply(input_image, model)