Prior to the bootcamp, we were assigned homework to set up PyRosetta and learn Python.
In homework #1, we install PyRosetta on our Google Colab Environments. It required some internal tweaking and a separate .whl from the developer. In its continuation, we check if everything works.
We then do homework #2 on Python for-loops, homework #3 on if-else statements, and homework #4 on functions.
Google Colab
To make everything work in Google Colab, I ended up having a PyRosetta directory with the .whl file in my Google Drive home directory MyDrive. Also, I kept the inputs, Media, and Sessions directories in a directory named temp_pyrbc_202103_notebooks.
We spent the first week of our internship at the University of North Carolina at Chapel Hill, where we learned PyRosetta and other useful skills that will come handy during the rest of our internships.
During the week, we worked on PyRosetta that was set up in Google Colab. Our teacher was Andrew Leaver-Fay (amazing guy, knows everything about Rosetta and the field).
We had around 15 sessions, starting with:
poseclass basics- PyMol visualizer
- Rosetta score functions like
scorefxn()
Getting into more advanced things:
- Movers to mutate residues
- Basic folding processes with algorithms like Centroid Folding
- XML scripts
- and many other things that you can find in the bootcamp directory.
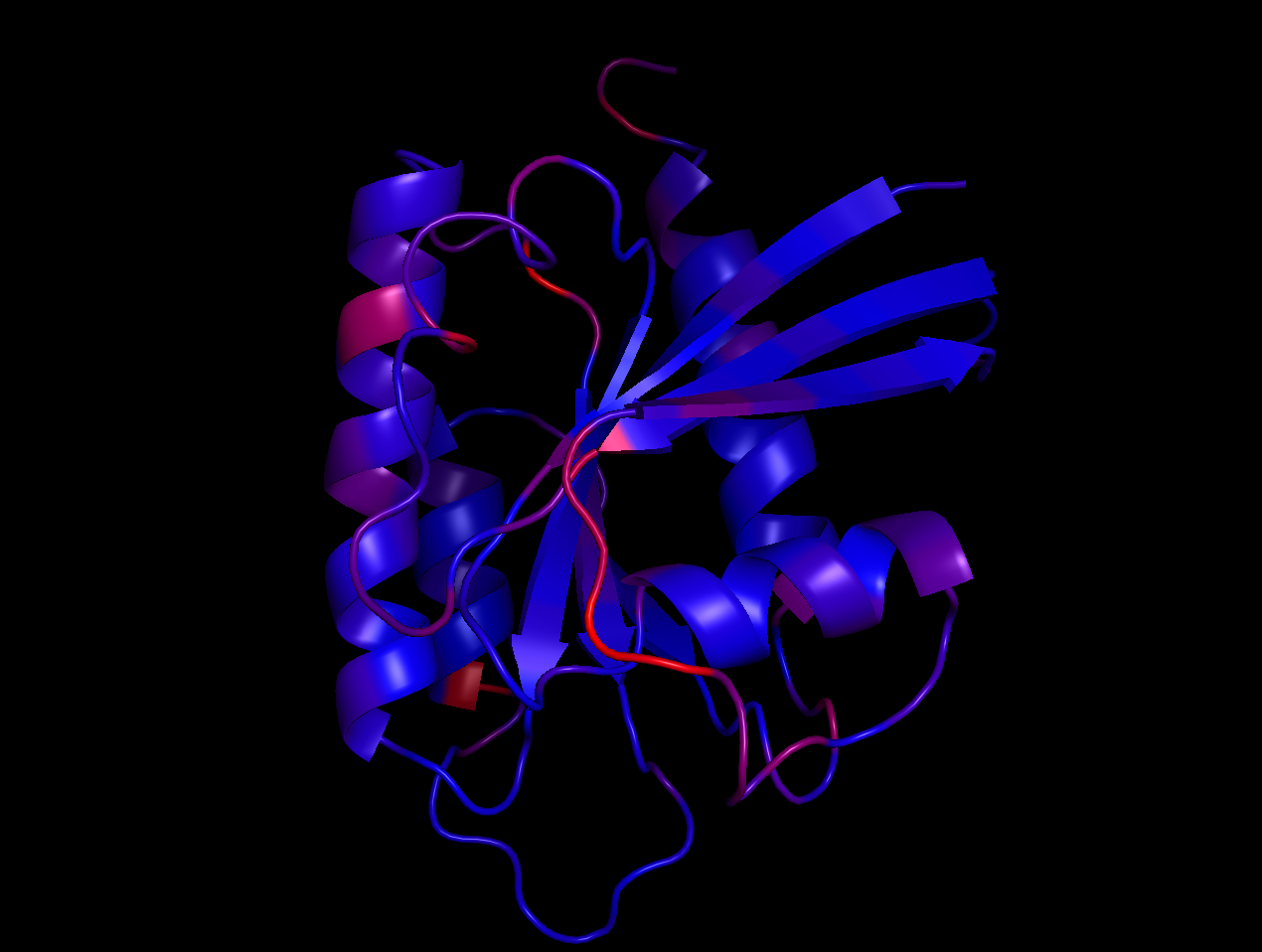
An example of what a protein fold looks like:
I've really enjoyed the bootcamp because I got to learn about a very exciting field of protein design and prediction, and I also got to meet amazing people from all over the US! You can learn more about my experience on my website.
During the rest of my internship, I worked at Stanford University, Possu Huang's Lab.
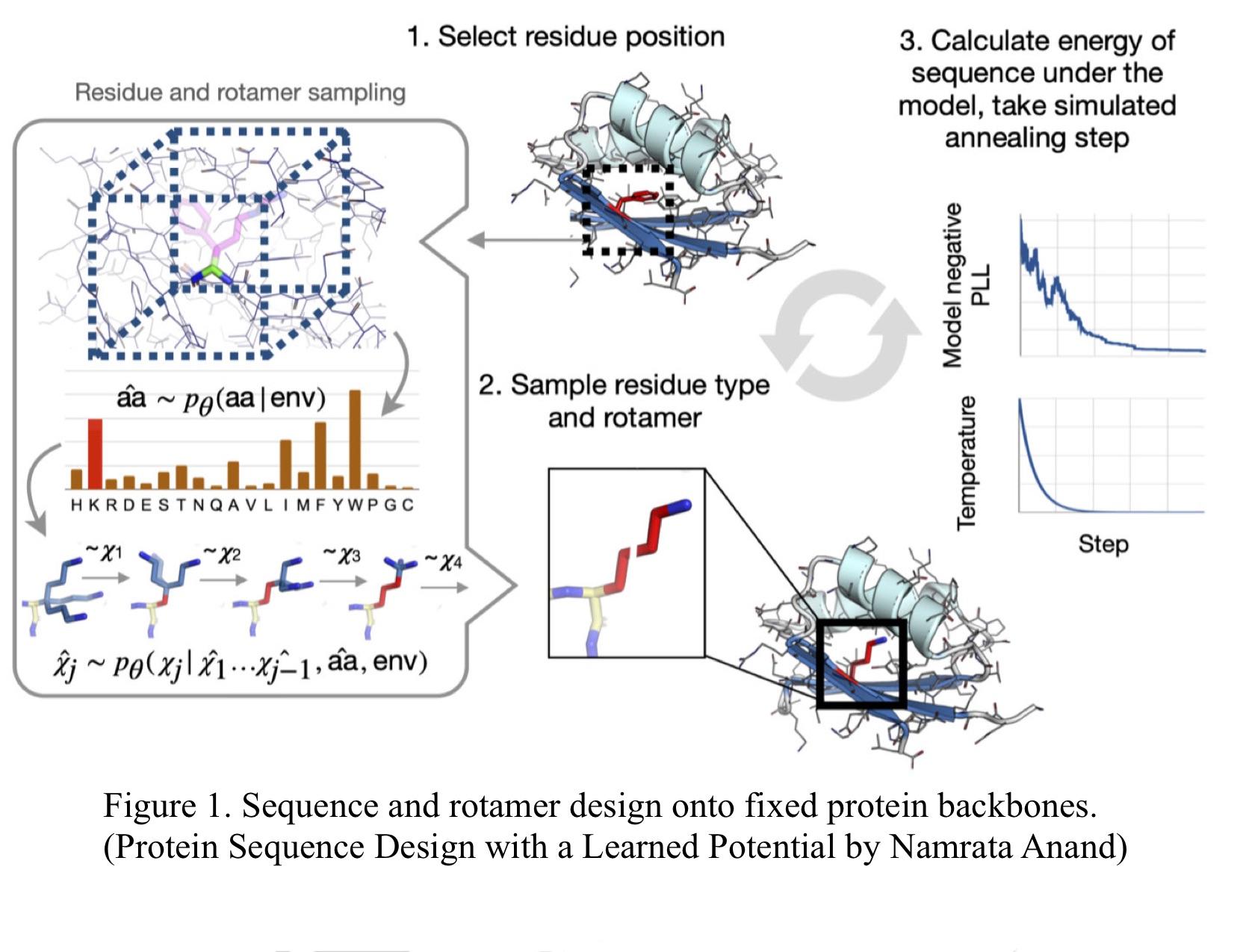
My research project is called Building Hydrogen Bonding Networks with Protein Sequence Design Model, for which I'm taking the Machine Learning model developed at the lab a step further. Their ML model, called Protein Sequence Design Model is a part of the algorithm that samples from the predicted distribution of long sequences of amino acids based on a fixed protein backbone for the inverse protein folding problem.
Based on the local environment, the model produces a distribution of possible residue types and rotamer angles (step 1). The algorithm then designs the protein through iterative sampling from the predicted distribution defined by the local chemical environment (step 2). The sampled residue type and rotamer angle values are then optimized using simulated annealing (step 3).