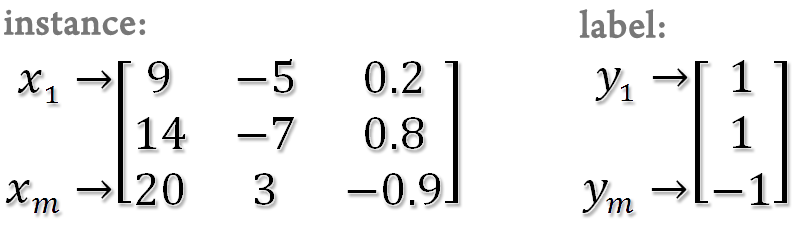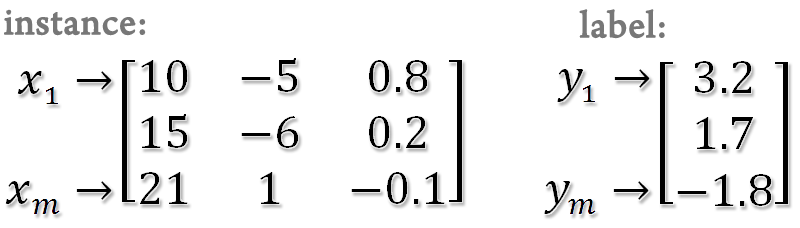SSVM toolbox is an implementation of Smooth Support Vector Machine in Matlab. SSVM is a reformulation of conventional SVM and can be solved by a fast Newton-Armijo algorithm. Besides, choosing a good parameter setting for a better performance in a learning task is an important issue. We also provide an automatic model selection tool to help users to get a good parameter setting. SSVM toolbox now includes smooth support vector machine for classification, epsilon-insensitive smooth support vector regression and an automatic model selection tool using uniform design.
- Solve classification (SSVM) and regression (SSVR) problems
- Support linear, polynomial and radial basis kernels
- Provide an automatic model selection for SSVM and SSVR with RBF kernels
- Can handle large scale problems by using reduced kernel (RSVM)
- Provide cross validation evaluation
- Provide an alternative initial point other than zero using regularized least squares
SSVM toolbox is implemented in Matlab. Use a data format which can be loaded into Matlab. The instances are represented by a matrix (rows for instances and columns for variables) and the labels (1 or -1) or responses are represented by a column vector.
For classification
For regression
Here are some sample datasets.
SSVM toolbox contains three main functions: ssvm_train for SVMs training, ssvm_predict for SVMs prediction and hibiscus for automatic model selection.
model = ssvm_train(label, inst, 'options')
| Variable | Description |
|---|---|
| label | training data class label or response |
| inst | training data inputs |
| Variable | Description |
|---|---|
| -s | learning algorithm (default: 0) 0-SSVM 1-SSVR |
| -t | kernel type (default: 2) 0-linear 1-polynomial 2-radial basis (Gaussian kernel) |
| -c | the weight parameter C of SVMs (default: 100) |
| -e | epsilon-insensitive value in epsilon-SVR (default: 0.1) |
| -g | gamma in kernel function (default: 0.1) |
| -d | degree of polynomial kernel function (default: 2) |
| -b | constant term of polynomial kernel function (default: 0) |
| -m | scalar factor of polynomial kernel function (default: 1) |
| -r | ratio of random subset size to the full data size (default: 1) |
| -i | alternatives initial point (default: 0) 0-using zero vector (w = 0, b = 0) 1-using an initial point obtained from regularized least squares |
| -v | number of cross-validation folds (default: 0) |
| Variable | Description |
|---|---|
| model | learning model (a structure in Matlab) |
| .w | normal vector of separating (or response) hyperplane |
| .b | bias term |
| .RS | reduced set |
| .Err | error rate (a structure in Matlab) |
| .Training | training error |
| .Validation | validation error |
| .Final | final model error in classification, it returns the error rate in regression, it returns the relative 2-norm error and the mean absolute error |
| .params | parameters specified by the user inputs |
- Classification using a Gaussian kernel
model = ssvm_train(label, inst, 'options') - Classification using a polynomial kernel and an initial point by regularized least squares
model_two = ssvm_train(label, inst, '-s 0 -t 1 -c 23.71 -d 2 -m 3 -b 2 -i 1') - SClassification using a Gaussian kernel and a 10% reduced set
model_three = ssvm_train(label, inst, '-s 0 -t 2 -c 421 -g 0.31 -r 0.1') - Classification using a Gaussian kernel in a 5-fold cross validation
model_four = ssvm_train(label, inst, '-s 0 -t 2 -c 23.71 -g 0.0625 -v 5') - Regression using the Gaussian kernel
model_five = ssvm_train(label, inst, '-s 1 -t 2 -c 10000 -g 0.71')
[PredictedLabel, ErrRate ]= ssvm_predict(label, inst, model)
| Variable | Description |
|---|---|
| label | testing data class label or response |
| inst | testing data inputs |
| model | model learned from ssvm_train or user-specified |
| Variable | Description |
|---|---|
| Predicted | Labelpredicted label or response |
| ErrRate | error rate (for classification) or relative 2-norm error (for regression) |
- prediction using model_one (classification problem)
[PredictedLabel, ErrRate]=ssvm_predict(label, inst, model_one)(Note: In this case, the error rate is meaningless.) - prediction using model_one without testing labels(classification problem)
[PredictedLabel, ErrRate]=ssvm_predict(zeros(NumOfInstm,1), inst, model_one) - prediction using model_five (regression problem)
[PredictedLabel, ErrRate]=ssvm_predict(label, inst, model_five)
Result = hibiscus(label, inst, 'method', 'type', 'command')
Note:only for Gaussian kernel
| Variable | Description |
|---|---|
| label | training data class label or response |
| inst | training data inputs |
| method | learning algorithm (default: 'SSVM') It must be {'SSVM', 'SSVR'} (case insensitive) |
| type | model selection methods (default: 'UD') It must be {'UD', 'GRID'} (case insensitive) |
| command | (optional) -v: number of cross-validation folds (default: 5) -r: ratio of random subset size to full data size (default: 1) |
| Variable | Description |
|---|---|
| Result | Include all returned information (a structure in Matlab) |
| .TErr | Training Error |
| .VErr | Validation Error |
| .Best_C | The best C in our model selection method |
| .Best_Gamma | The best gamma in our model selection method |
| .Elapse | CPU time in seconds |
| .Points | Trying points |
| .Ratio | Ratio of random subset size to full data size |
- Determine C and Gamma by a 5-fold cross-validation and the full kernel for classification
Result = hibiscus(label, inst, 'SSVM', 'UD', '-v 5 -r 1') - Determine C and Gamma by a 10-fold cross-validation and a 10% reduced kernel for classification
Result = hibiscus(label, inst, 'SSVM', 'UD', '-v 10 -r 0.1') - Determine C and Gamma by a 10-fold cross-validation and 10% reduced kernel for regression
Result = hibiscus(label, inst, 'SSVR', 'UD', '-v 10 -r 0.1')
- Change your current directory to SSVMtoolbox folder
- Load dataset from Ionosphere_dataset.mat (can be found in SSVMtoolbox)
load Ionosphere_dataset.mat
- Determine good C and gamma via a 5-fold cross validation using full kernel
Result = hibiscus(label, inst, 'SSVM', 'UD', '-v 5 -r 1');
- Read the contents of Result (Best_C, Best_Gamma, ...etc)
Result
Result.Best_C
- Training a model by using Best_C and Best_Gamma in Result
model = ssvm_train(label, inst, '-s 0 -t 2 -c 10 -g 0.0856');
- Read the contents of model (w, b, parameters, ...etc)
model
model.w
- Prediction using the model obtained from the training process (here, we use the training set as the testing set)
[PredictedLabel, ErrRate]=ssvm_predict(label, inst, model);
- Change your current directory to SSVMtoolbox folder
- Load dataset from housing_dataset.txt (can be found in SSVMtoolbox)
load housing_dataset.txt
- Split the housing data into inst and label
inst = housing_dataset(:,1:13);
label = housing_dataset(:,14);
- Determine good C and gamma via a 5-fold cross validation and 10% reduced kernel
Result = hibiscus(label, inst, 'SSVR', 'UD', '-v 5 -r 0.1');
- Read the contents of Result (Best_C, Best_Gamma, ...etc)
Result
Result.Best_C
- Training a model by using Best_C and Best_Gamma in Result
model=ssvm_train(label, inst, '-s 1 -c 1000000 -g 0.1794 -r 0.1');
- Read the contents of model (w, b, parameters, ...etc)
model
model.w
- Prediction using the model obtained from the training process (here, we use the training set as the testing set)
[PredictedLabel, ErrRate]=ssvm_predict(label, inst, model);
- Read the value of relative 2-norm error and mean absolute error
ErrRate(1)
ErrRate(2)
This software is available for non-commercial use only. The authors are not responsible for implications from the use of this software.