HORDE
- This is a Tensorflow implementation of "Harmonized Representation Learning on Dynamic EHR Graphs", published in Journal of Biomedical Informatics 2020.
- The PyTorch implementation is available at https://github.com/Lishany/HORDE-pytorch.
Overview
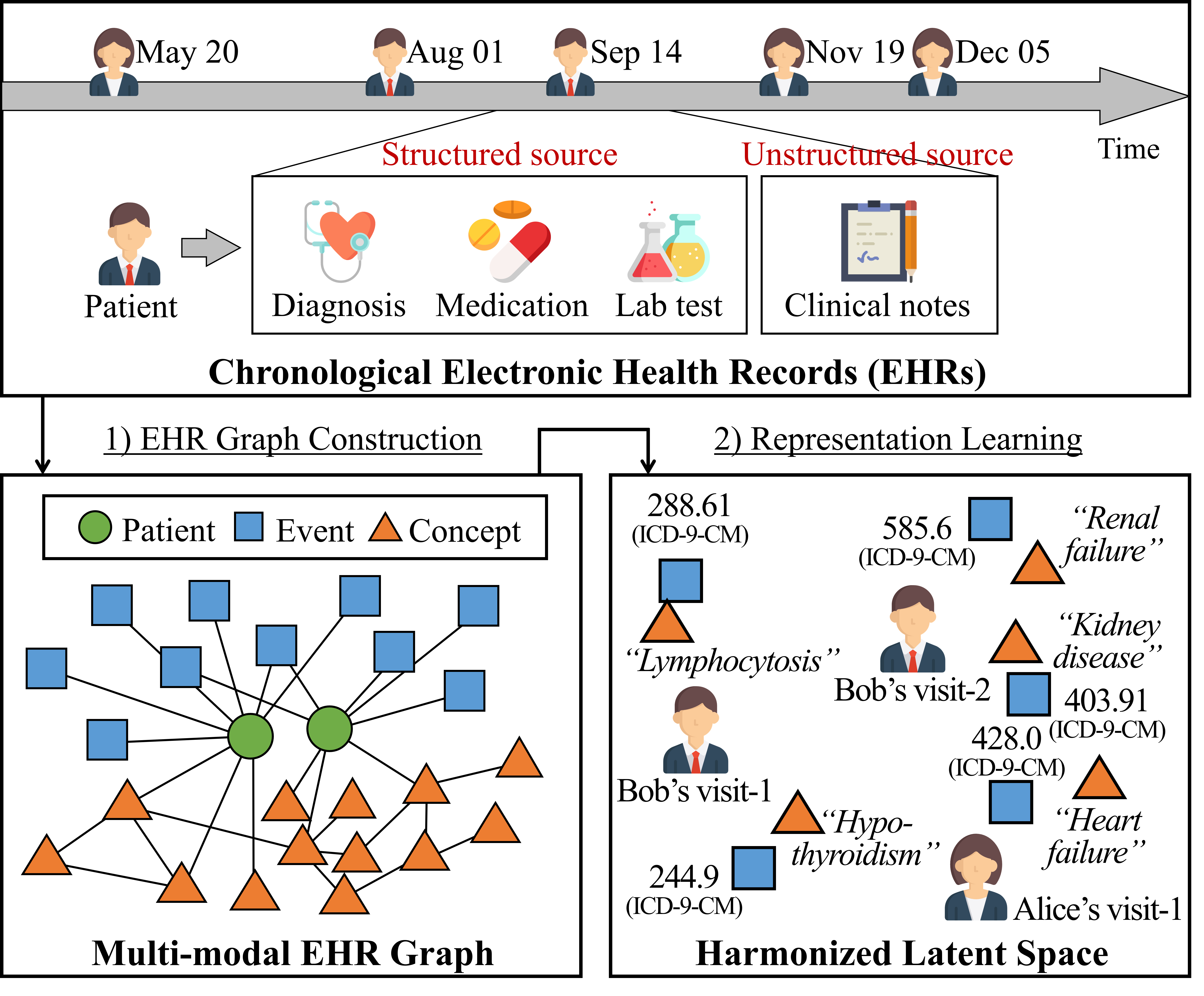
The overview of harmonized representation learning on the EHR. The overall process consists of two parts: 1) multi-modal EHR graph construction, and 2) graph representation learning.
Running the Code
Step 1. Installing Python and Tensorflow
- Install
Python 3. (numpy, scipy, and sklearn packages are required as well.) - Install
tensorflow-gpudocker. You can easily pull it from the NVIDIA cloud by the following pull command:
docker pull nvcr.io/nvidia/tensorflow:19.01-py3
- Please use tensorflow 1.x, or modifiy the codes so that it can be running on tensorflow >= 2.0.
Step 2. Preparing Data
- You need .npy (numpy file format) file of EHR.
- It should be in the form of numpy array, which includes the list of visits.
- Each visit has to contain
patient id,visit id,diagnosis code list, andNLP concept list.- For example,
[[3, 1, ['460', '460.1'], ['Anorexia', 'Hypertensive disease']],
[3, 2, ['V70.0', '625.9'], ['Hepatitis', 'Hepatitis', 'Vaginitis']],
...,
[3243123, 5, ['512.1', '518.0'], ['Atelectasis', 'Pneumonia', 'Pneumothorax']]]
- You do not need to enforce visit-ids unique. They do not matter to run the codes.
- Multiple visits of a single patient do not have to be consecutive, but they should be sorted in a chronological order.
- Diagnosis codes and NLP concepts must be provided as string format.
- NLP concepts should preserve their order in original clinical notes, and they can occur multiple times in the list.
- Run the script for data preprocessing by the following command:
python process_data.py <input_numpy_file> <output_directory> <test_patients_number>
- Test patients are used to evaluate the representation quality.
- Then, three files would be created in the output directory;
graph.npy,ctxpairs.npy, andpatients.npy.
Step 3. Running HORDE on your Data
Now, you are ready to learn the representations of medical entities based on HORDE.
- if you want to use CPU
python main.py --input_dir <input_directory> --output_dir <output_directory>
- if you want to use GPU (GPU:0 is used unless gpu_devidx is specified)
python main.py --gpu True --gpu_devidx <GPU_device_index> --input_dir <input_directory> --output_dir <output_directory>
These are additional possible arguments you can specify:
| Argument | Description |
|---|---|
| --input_dir | The path to the input nummpy files (the output directory at the step 2) |
| --output_dir | The path to the output embedding numpy files |
| --vector_size | The size of the final representation vectors (default: 256) |
| --negsample_size | The number of negative context pairs per positive context pair (default: 1) |
| --learning_rate | The initial learning rate for the ADMM optimizer (default: 0.001) |
| --ti_dropout | The dropout rate for time-invariant node vectors (default: 0.3) |
| --tv_dropout | The dropout rate for time-variant node vectors (default: 0.3) |
| --ti_batch_size | The size of a single mini-batch for time-invariant node pairs (default: 512) |
| --tv_batch_size | The size of a single mini-batch for time-variant node sequences (default: 32) |
| --weight_decay | The coefficient of L2 regularization on all the weight parameters (default: 0.001) |
| --n_iters | The total number of mini-batches for training (default: 200,000) |
| --n_printiters | The number of mini-batches for evaluating the model and printing outputs (default: 2,000) |
| --recall_at | The k value in Recall@k for subsequent event prediction (default: 30) |
Step 4. Tuning the Hyperparameters
We strongly recommend to tune the following hyperparameters, whose combination affects the quality of representations.
- --ti_batch_size ∈ {128, 256, 512}
- --tv_batch_size ∈ {16, 32, 64}
EHR Analysis
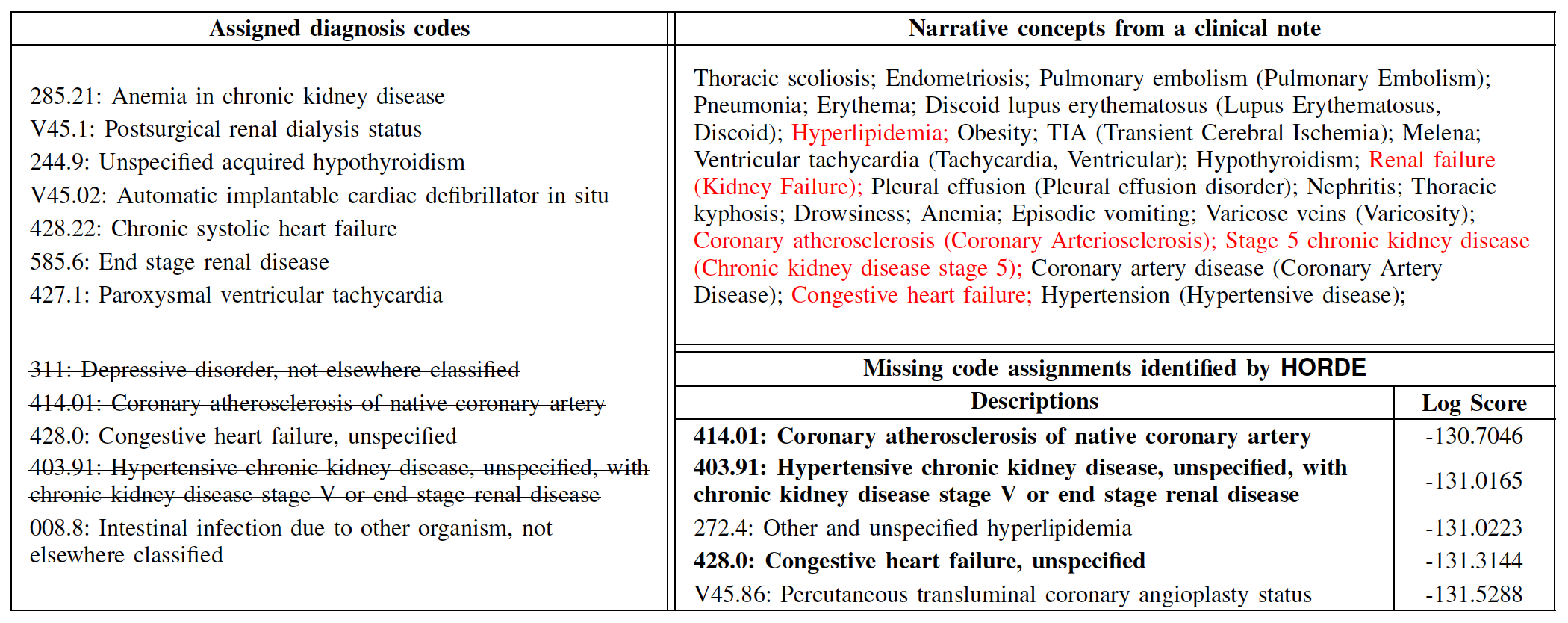
- Using the representations obtained by
HORDE:- we can identify less consistent code assignments (i.e., more likely to be erroneous) from the visits of a patient.
- we can infer missing codes for each visit by retrieving the codes that are not assigned but have high scores.
A detailed example of our consistency analysis on deletion-type noises (N = 5, MIMIC-III). The left column is the list of assigned codes in a target visit, and strike-through texts represent randomly deleted code assignments. The upper part of the right column shows all narrative concepts extracted from a discharge summary of the visit, and the lower part is the identification result of N missing codes by the highest confidence scores from
HORDE.
Citation
Dongha Lee, Xiaoqian Jiang, and Hwanjo Yu. "Harmonized Representation Learning on Dynamic EHR Graphs", Joural of Biomedical Informatics, 2020.
@article {lee2020harmonized,
title = {Harmonized representation learning on dynamic EHR graphs},
author = {Lee, Dongha and Jiang, Xiaoqian and Yu, Hwanjo},
volume = {106},
year = {2020},
journal = {Journal of biomedical informatics},
pages = {103426}
}