- This is a PyTorch implementation of "Generating sequential electronic health records using dual adversarial autoencoder", published in Journal of the American Medical Informatics Association (JAMIA) 2020.
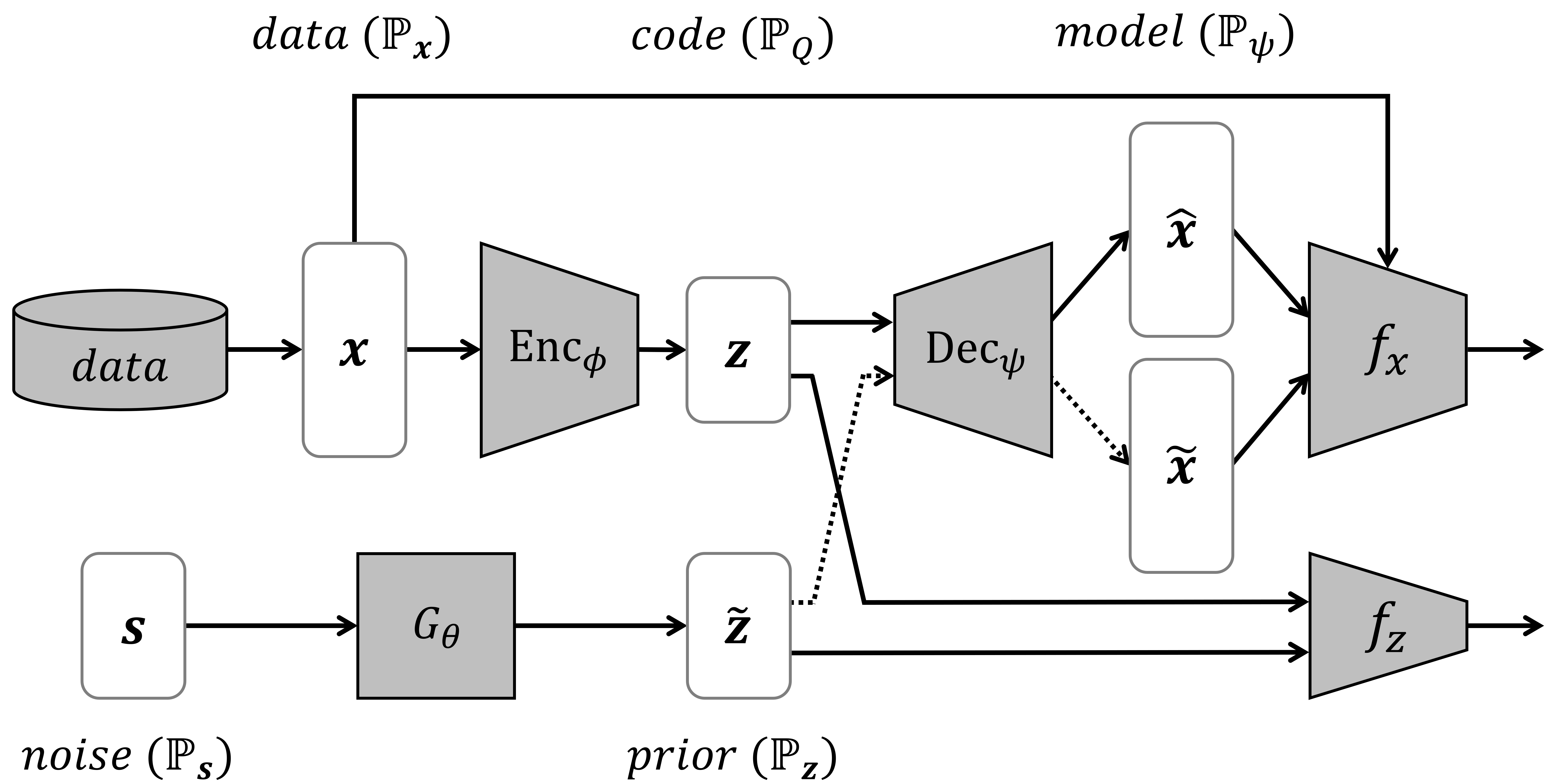
The DAAE architecture that is composed of 1) the seq2seq autoencoder, 2) the inner GAN, and 3) the outer GAN.
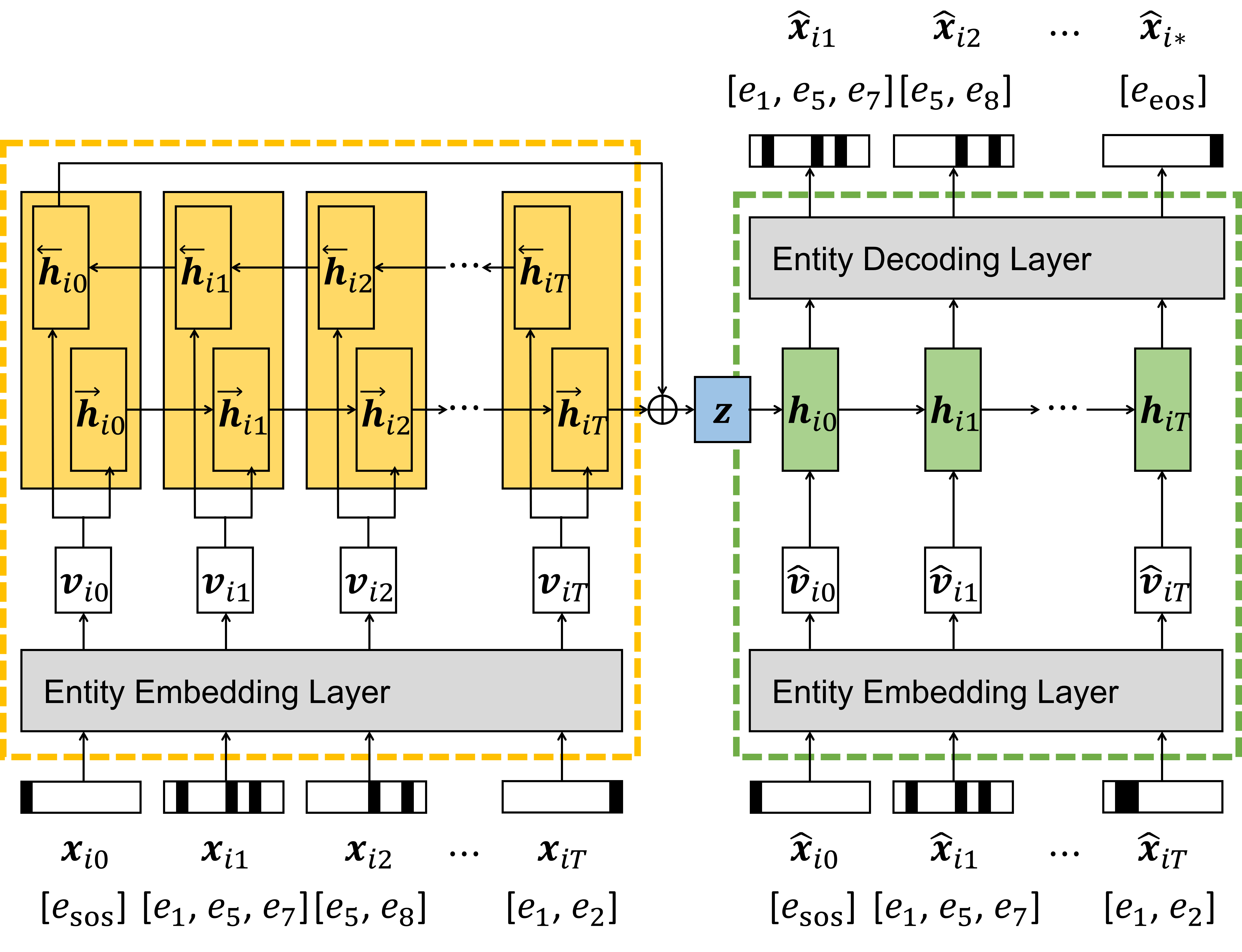
The detailed architecture of the seq2seq autoencoder. The embedding layer encodes the semantics of all the medical entities, and the GRU layer learns the temporal contexts within patients' sequential records.
- Install
Python 3andPyTorch.
- You need .npy (numpy file format) files of EHR.
- It should be in the form of a dictionary, whose
keyis the patient-id andvalueis the set-valued sequence of the patient. - For example,
{ 17: [['7455', '45829', 'V1259', '2724'], ['4239', '5119', '78551', '4589', '311', '7220', '71946', '2724']],
21: [['41071', '78551', '42731', '1122', '2720', '2749', 'V1046', '43889'], ['0388', '78552', '40391', '42731', '25000', '2859', '43889', '2749', '41401', '185', '4439', '2449']],
...,
99982: [['42823', '4254', '2875', '5303', '4280', 'V5861', '45829'], ['4280', '42823', '5849', '4254', '2763', '42731', '78729', '2768'], ['5849', '42731', '4280', '2875','V422', '7994']]}
- Each entity (i.e., diagnosis code) must be provided as string format.
- In the input data directory, two .npy files should be required respecitvely for training and validation:
ehr.train.npyandehr.valid.npy.
- Now, you are ready to learn the sequences about real patients and synthesize fake ones.
python train_daae.py --data_dir <input_directory> --create_data --gendata_size <#_fake_sequences>
- Finally, you will obtain the two files about fake patients:
daae_generated_codes.npyanddaae_generated_patients.npy. - These are basic arguments you can specify:
| Argument | Description |
|---|---|
| --data_dir | The path to the input nummpy files |
| --max_sequence_length | The maximum length of sequential records (default: 10) |
| --max_visit_length | The maximum number of entities in a single record (default: 40) |
| --gendata_size | The number of fake sequences to be generated (default: 100,000) |
| --batch_size | The size of a single mini-batch for set-valued sequences (default: 32) |
| --learning_rate | The initial learning rate for the ADMM optimizer (default: 0.001) |
| --epochs | The total number of epochs for training (default: 500) |
| --gpu_devidx | The index of GPU to be used (default: 0) |
- The detailed architecture of each componenet (i.e., encoder, decoder, generator, and inner/outer critics) can be specified by using the following arguments:
| Argument | Description |
|---|---|
| --rnn_type | The type of RNN encoder and decoder (default: 'gru') |
| --embedding_size | The dimensionality of the entity embedding layer (default: 128) |
| --hidden_size | The dimensionality of hidden vectors in RNN encoder and decoder (default: 128) |
| --noise_size | The dimensionality of noise vectors (default: 128) |
| --latent_size | The dimensionality of code vectors (default: 128) |
| --filter_size | The number of conv filters in the outer critic (default: 16) |
| --window_sizes | The widths of conv filters in the outer critic (default: [2, 3]) |
| --bidirectional | The flag indicating the RNN encoder is bidirectional (default: False) |
| --entity_dropout | The entity dropout rate for decoder input sequences (default: 0.05) |
| --embedding_dropout | The dropout rate for the entity embedding layer (default: 0.5) |
| --feature_dropout | The dropout rate for the fc layer in the outer critic (default: 0.5) |
| --gmlp_archs | The MLP architecture of the generator (default: [128, 128]) |
| --dmlp_archs | The MLP architecture of the inner critic (default: [256, 128]) |
- If you need to guarantee the synthetic samples do not leak private information about the training data, you can use a DP optimizer when training the model.
- Install the PyVacy package and run the code with
--dp_sgdset toTrue. - You can control the privacy budget by the following arguments:
--noise_multiplier,--l2_norm_clip, and--delta. - Note that there is a trade-off between the privacy guarantee and the quality of generated sequences.
@article{lee2020generating,
author = {Lee, Dongha and Yu, Hwanjo and Jiang, Xiaoqian and Rogith, Deevakar and Gudala, Meghana and Tejani, Mubeen and Zhang, Qiuchen and Xiong, Li},
title = {Generating sequential electronic health records using dual adversarial autoencoder},
journal = {Journal of the American Medical Informatics Association},
volume = {27},
number = {9},
pages = {1411-1419},
year = {2020},
month = {09}
}