This project aims to conduct a comprehensive analysis of the impact of the Covid-19 pandemic on Germany. Utilizing geographic and COVID-19 data sourced from the World Health Organization (WHO), the project attempts to offer an understanding of the virus' spread and its correlation with healthcare indicators. The primary objective is to analyze the relationship between total cases, population numbers, and death rates. By doing so, the project seeks to provide valuable insights into the effectiveness of healthcare systems across different regions in Germany in managing and mitigating the impact of future pandemics.
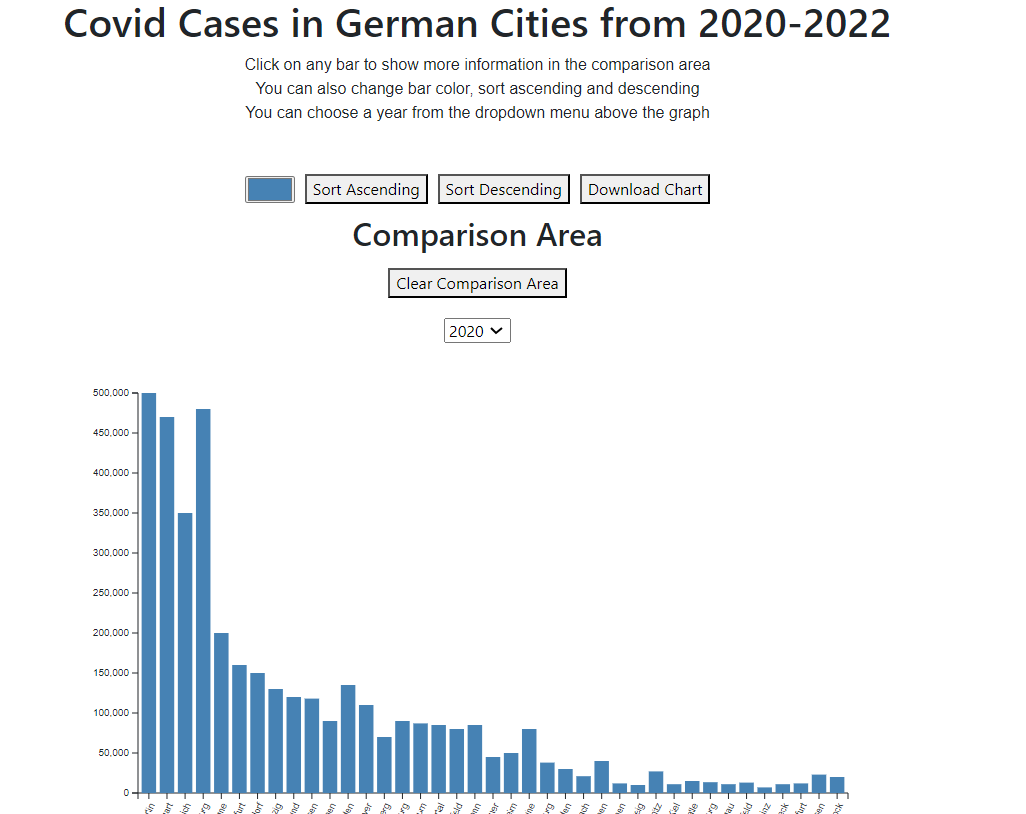
Insights for each year: Covid data visualization for covid cases throughout each year of the pandemic in the 40 most populated German cities.
This project leverages several key technologies and frameworks to provide a robust and interactive data visualization experience:
- Python: The primary backend language, known for its readability and wide support in data processing and web development.
- FastAPI: A modern, high-performance web framework for Python, used for creating efficient API endpoints.
- SQLite: A lightweight database to store and manage the COVID-19 data in a structured format.
- D3.js: A powerful JavaScript library for producing dynamic, interactive data visualizations in web browsers.
- HTML/CSS/JavaScript: The core technologies of the web for structuring, styling, and adding interactivity to the web pages.
- JSON: Used for data storage and transfer, particularly for uploading new datasets and exchanging data between the backend and frontend.
Public Health Researchers: This tool equips researchers with detailed, city-specific COVID-19 data, aiding in analyzing trends, preparing public health strategies, and evaluating policy effectiveness. The comprehensive data and visualizations facilitate deeper insights into the pandemic's dynamics, essential for shaping future public health initiatives.
The core functionality of this project lies in providing a dynamic and comparative analysis of COVID-19 data across various cities in Germany, which is pivotal for identifying patterns, determining hotspots, and recognizing anomalies in the virus's transmission. In a real-world context, this feature empowers researchers to leverage city-specific insights for investigating correlations between variables such as population density and virus spread. This kind of analysis is invaluable for future urban planning and healthcare resource management, helping to strategize more effectively against current and future public health challenges.
Scenario 1: Analyzing Yearly COVID-19 Trends in Major Cities
Researchers can select individual years (2020, 2021, 2022) to visualize the trend of COVID-19 cases in the most populated cities of Germany. This feature allows for a comparative analysis of how the pandemic evolved over the years.
Scenario 2: Evaluating the Impact of Population Density
Public health officials can use the tool to correlate population density with COVID-19 spread in different cities. This insight is crucial for resource allocation and planning for future healthcare emergencies.
Scenario 3: Assessing Vaccination Impact
By comparing the vaccination rates with the number of COVID-19 cases and deaths, healthcare policymakers can gauge the effectiveness of vaccination campaigns and strategize accordingly for future public health initiatives.
-
Open in GitHub Codespaces:
- Go to the GitHub repository: [https://github.com/ali-b7/Covid-Analysis-in-Germany.git].
- Click on 'Code', then select 'Open with Codespaces'.
- Choose 'New codespace' to create a new development environment.
-
Automatic Setup:
- Once open in GitHub Codespaces, the development container will automatically set up the environment for you.
- However, if that does not work then run
uvicorn main:app --host 127.0.0.1 --port 8000 --reloadin the terminal.
-
Accessing the Application:
- Once the Codespace environment is ready and the application starts, go to the 'Ports' panel.
- Find the port
8000listed there. - Click on 'Open in Browser' next to port
8000. This will open a new browser tab or window with the application running.
-
Clone the Repository:
- In the terminal, execute:
git clone https://github.com/ali-b7/Covid-Analysis-in-Germany.git - Navigate to the cloned directory:
cd Covid-Analysis-in-Germany
- In the terminal, execute:
-
Set Up a Virtual Environment (Optional but Recommended):
- Create a virtual environment:
python -m venv env - Activate the environment:
- Windows:
env\Scripts\activate - macOS/Linux:
source env/bin/activate
- Windows:
- Create a virtual environment:
-
Install Dependencies:
- Ensure your virtual environment is activated.
- Install required packages:
pip install -r requirements.txt
-
Starting the Application:
- Start the FastAPI server:
uvicorn main:app --host 127.0.0.1 --port 8000 --reload - The application will be accessible at
http://localhost:8000. - Uploading data works the same way as in running the applicaiton in a codespaces.
- Start the FastAPI server:
- If you encounter any issues with package installation, try upgrading pip:
pip install --upgrade pipand rerun the requirements installation. - Ensure that the ports specified (e.g., 8000) are not in use. If they are, either free up the port or specify a different port in the
uvicorncommand.
- Above the graph you can find the option to upload your test json file or visit the route /docs.
-
After being redirected, click on the green "POST" link to show the route /uploadjsondata .Click on try it out and choose your file which must be based on the sample json data provided either in the repository or at the upload location of the examination.
-
Click on execute and see if the upload worked (status 200) or if it was rejected because of format or wrong file type (status 500)
-
Delete the /docs so that you only have the regular address of your codespace port see the uploaded data along with my original files in the visualisation.
-
If you want to return to the original data then terminate the app in the terminal and restart it.
Contributions are welcomed from the community! If you're interested in enhancing the capabilities of this tool or have innovative ideas for new features, here’s how you can contribute:
- Fork the Repository: Start by forking the repository to make your own version.
- Create a Pull Request: After making your changes, create a pull request. Please provide a clear description of what your changes achieve.
- Code Reviews: Your pull request will be reviewed. This is a space for discussion and improvements.
- Merge: Once approved, your contributions will be merged into the main project.
For support, feedback, or contributions, feel free to contact me:
- Email: ali.badran@stud.th-deg.de