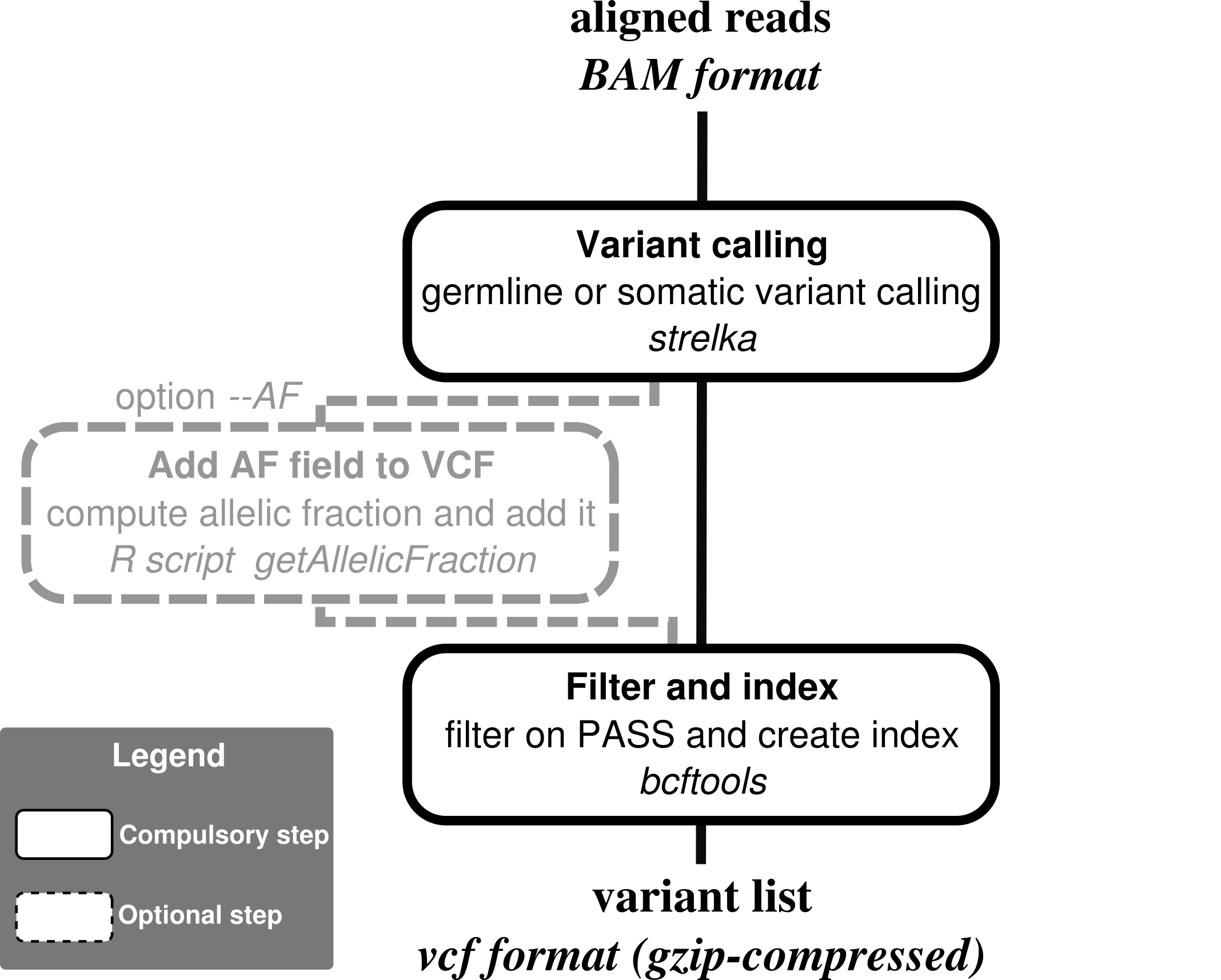
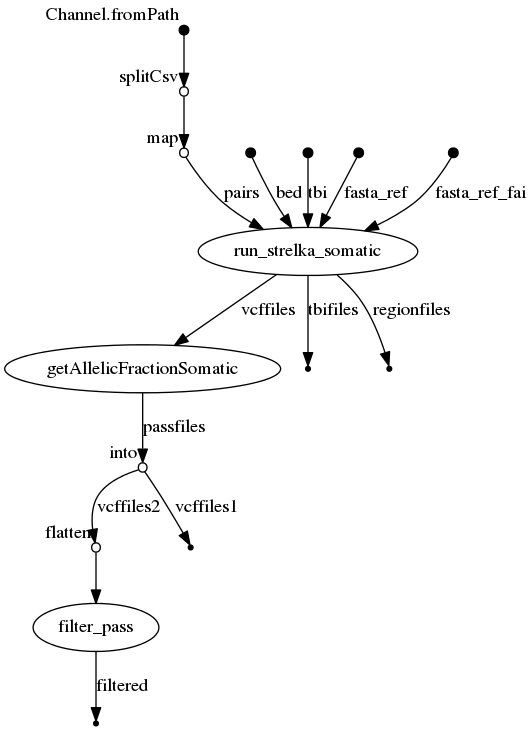
- Nextflow : for common installation procedures see the IARC-nf repository.
- Install Strelka v2.
| Type | Description |
|---|---|
| --input_folder | folder with bam/cram files |
| --input_file | Tab delimited text file with either two columns called normal and tumor (somatic mode) or one column called bam (germline mode); optionally, a column called sample containing sample names to be used for naming the files can be provided and for genotyping (see genotyping mode below) a column called vcf has to be provided |
Note: the file provided to --input_file is where you can define pairs of bam/cram to analyse with strelka in somatic mode. It's a tabular file with 2 columns normal and tumor.
| normal | tumor |
|---|---|
| normal1.cram | tumor2.cram |
| normal2.cram | tumor2.cram |
| normal3.cram | tumor3.cram |
| Name | Example value | Description |
|---|---|---|
| --ref | hg19.fasta | genome reference |
| Name | Default value | Description |
|---|---|---|
| --mode | somatic | Mode for variant calling; one of somatic, germline, genotyping |
| --output_folder | strelka_ouptut | Output folder for vcf files |
| --cpu | 2 | number of CPUs |
| --mem | 20 | memory |
| --strelka | path inside docker and singularity containers | Strelka installation dir |
| --config | default conf of strelka | Use custom configuration file |
| --callRegions | none | Region bed file |
| --ext | cram | extension of alignment files (bam or cram) |
Flags are special parameters without value.
| Name | Description |
|---|---|
| --help | print usage and optional parameters |
| --exome | automatically set up parameters for exome data |
| --rna | automatically set up parameters for rna data (only available for --mode germline) |
| --AF | Add AF field to VCF (only available for --mode somatic) |
| --outputCallableRegions | Create a BED track containing regions which are determined to be callable |
nextflow run iarcbioinfo/strelka2-nf r v1.2a -profile singularity --mode somatic --ref hg38.fa --tn_pairs pairs.txt --input_folder path/to/cram/ --strelka path/to/strelka/
To run the pipeline without singularity just remove "-profile singularity". Alternatively, one can run the pipeline using a docker container (-profile docker) the conda receipe containing all required dependencies (-profile conda).
nextflow run iarcbioinfo/strelka2-nf r v1.2a -profile singularity --mode germline --ref hg38.fa --input_folder path/to/cram/ --strelka path/to/strelka/
When using the input_file mode, if a vcf column with the path to a VCF file for each sample containing a list of somatic variant is provided, the pipeline will use the --forcedGT option from strelka that genotypes these positions, and compute a bedfile for these positions so only variants from the VCF will be genotyped. Note that genotyping can be performed both in somatic mode (in which case tumor/normal pairs must be provided) and germline mode (in which case a single cram file must be provided).
| Type | Description |
|---|---|
| VCFs/raw/*.vcf.gz | VCF files before filtering |
| VCFs/withAF/*.vcf | VCF files with AF field (optional, requires flag --AF) |
| VCFs/filtered/*PASS.vcf.gz | final compressed and indexed VCF files (optionally with flag --AF) |
| CallableRegions/*.bed.gz | compressed and indexed BED files (optionally with flag --outputCallableRegions) |
Final vcf files have companion tabix index files (.tbi). Note that in germline mode, the VCF outputted corresponds to variants only (file variants.vcf.gz from strelka).
| Name | Description | |
|---|---|---|
| Vincent Cahais | CahaisV@iarc.fr | Developer |
| Nicolas Alcala | AlcalaN@iarc.fr | Developer |