cff-validator
A GitHub action to validate CITATION.cff files with R.
Introduction
If you have a Citation File Format (cff) on your repository this action would check its validity against the defined schema.
A full valid workflow:
on:
push:
paths:
- CITATION.cff
workflow_dispatch:
name: CITATION.cff
jobs:
Validate-CITATION-cff:
runs-on: ubuntu-latest
name: Validate CITATION.cff
env:
GITHUB_PAT: ${{ secrets.GITHUB_TOKEN }}
RSPM: "https://packagemanager.rstudio.com/cran/__linux__/focal/latest"
steps:
- name: Checkout
uses: actions/checkout@v2
# This is needed for workflows running on
# ubuntu-20.04 or later
- name: Install V8
if: runner.os == 'Linux'
run: |
sudo apt-get install -y libv8-dev
- name: Validate CITATION.cff
uses: dieghernan/cff-validator@main
# Upload artifact
- uses: actions/upload-artifact@v2
if: failure()
with:
name: citation-cff-errors
path: citation_cff_errors.mdOn error, the action shows the results of the validation highlighting the fields with errors.
It also generates an artifact named
citation-cff-errors that includes a
markdown file with a
high-level summary of the errors found:
citation_cff_errors.md
Table: **./examples/key-error/CITATION.cff errors:**| field | message |
|---|---|
| data | has additional properties |
| data.authors.0 | no schemas match |
| data.doi | referenced schema does not match |
| data.keywords.0 | is the wrong type |
| data.license | referenced schema does not match |
| data.url | referenced schema does not match |
See Guide to Citation File Format schema version 1.2.0 for debugging.
For more examples, see the actions provided on this path.
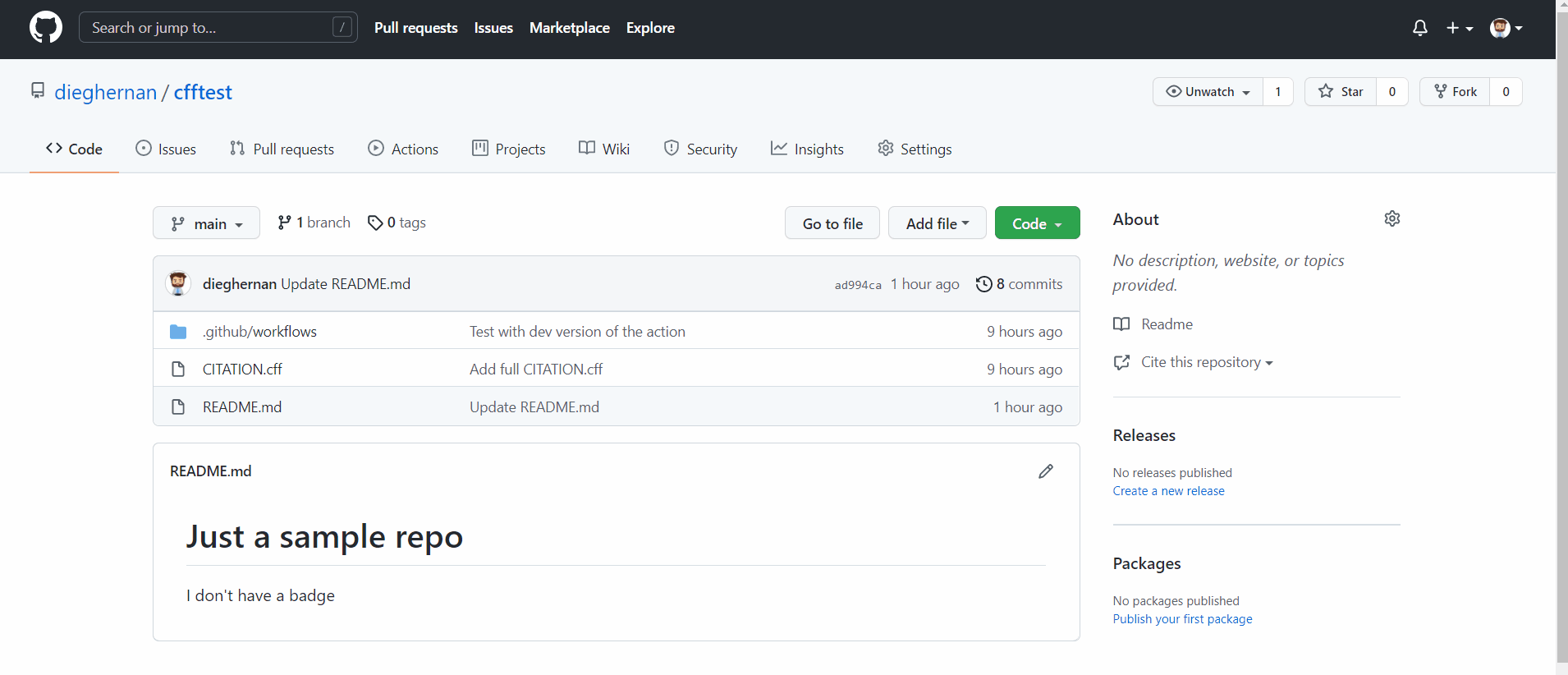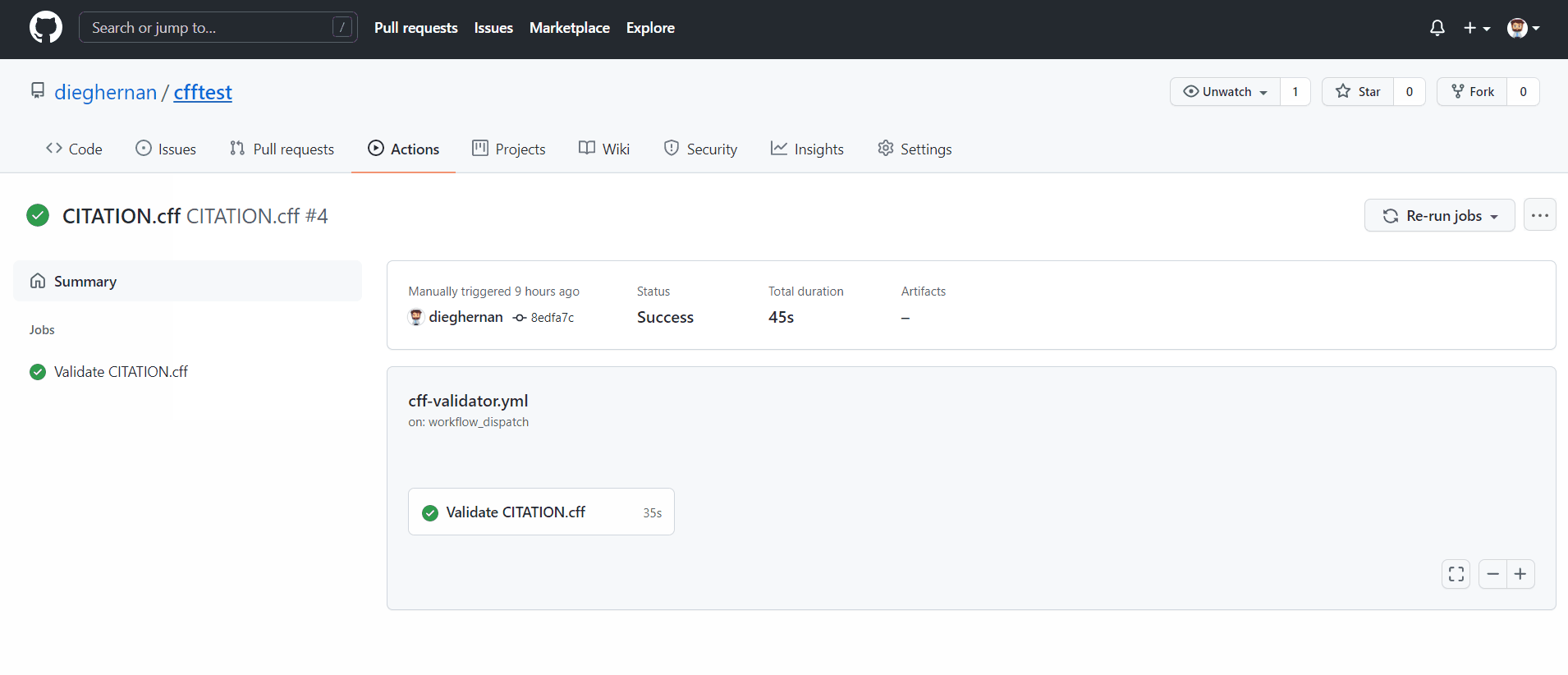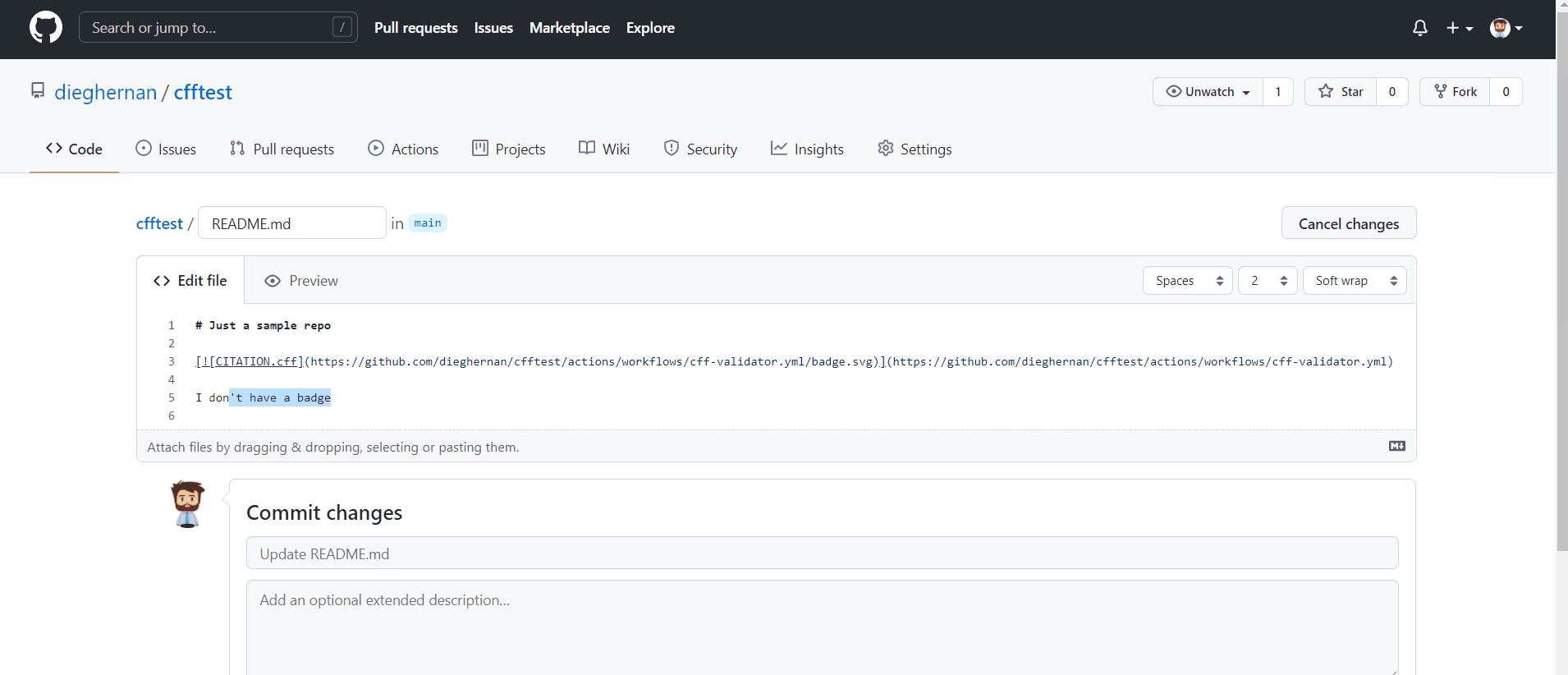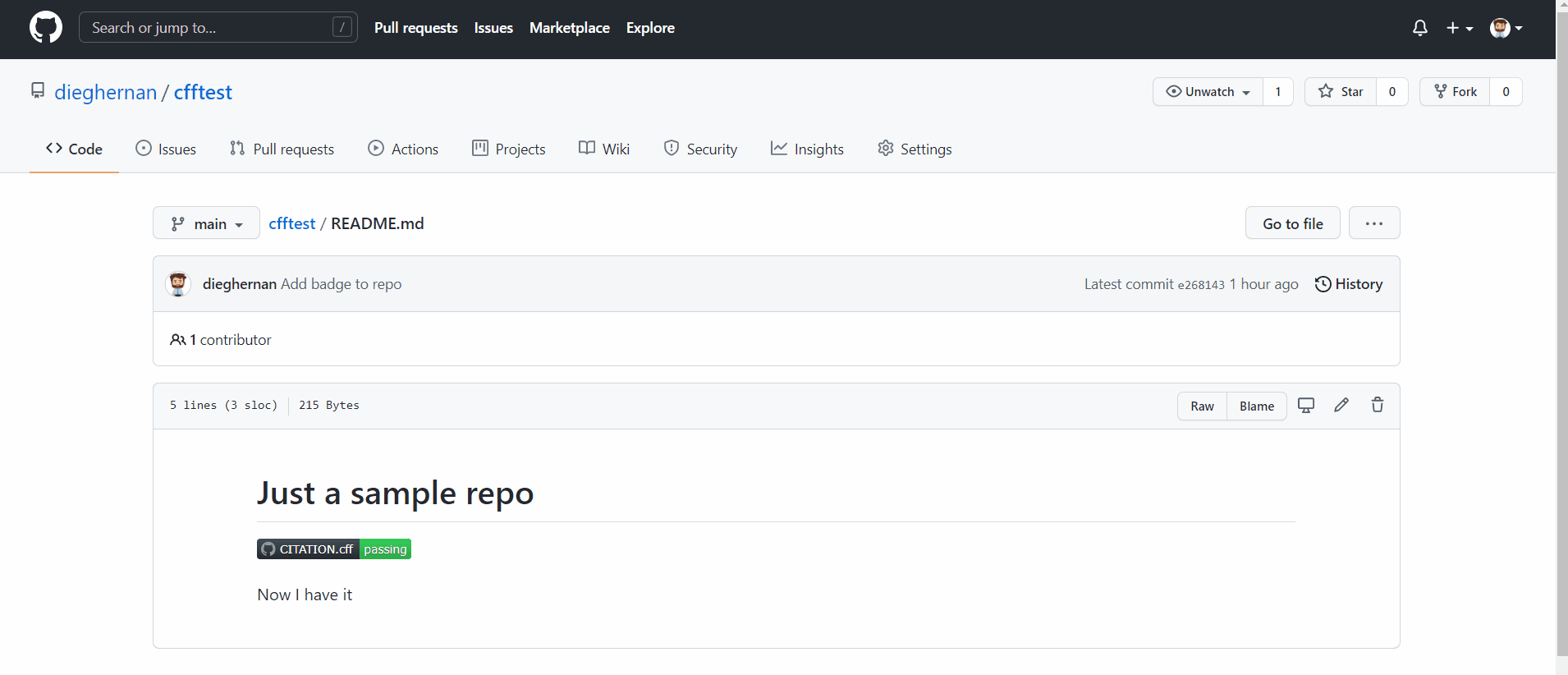
Add a badge to your repo
You can easily create a badge showing the current status of validation of your CITATION.cff like this:
See a quick demo:
Inputs available
citation-path: Path to .cff file to be validated. By default it selects aCITATION.cfffile on the root of the repository:
- name: Validate CITATION.cff
uses: dieghernan/cff-validator@main
with:
citation-path: "examples/CITATION.cff"Building on Linux
This action relies on the R package V8, that has some extra requirements when running on Linux systems. You would need to add the following steps to your action in order to make it run:
# This is needed for workflows running on
# ubuntu-20.04 or later
- name: Install V8
run: |
sudo apt-get install -y libv8-dev
# This is needed for workflows running on
# previous versions of ubuntu
- name: Install V8 on old ubuntu
run: |
# Ubuntu Xenial (16.04) and Bionic (18.04) only
sudo add-apt-repository ppa:cran/v8
sudo apt-get update
sudo apt-get install libnode-devSee a full featured implementation on this example.
Under the hood (for useRs)
This action runs a R script that can be easily replicated. See a full reprex:
R script
# install_cran(c("yaml","jsonlite", "jsonvalidate", "knitr")
citation_path <- "./key-error/CITATION.cff"
citfile <- yaml::read_yaml(citation_path)
# All elements to character
citfile <- rapply(citfile, function(x) as.character(x), how = "replace")
# Convert to json
cit_temp <- tempfile(fileext = ".json")
jsonlite::write_json(citfile, cit_temp, pretty = TRUE)
# Manage brackets
citfile_clean <- readLines(cit_temp)
# Search brackets to keep
# Keep ending and starting
keep_lines <- grep('", "', citfile_clean)
keep_lines <- c(keep_lines, grep("\\[$", citfile_clean))
keep_lines <- c(keep_lines, grep(" \\],", citfile_clean))
keep_lines <- c(keep_lines, grep(" \\]$", citfile_clean))
keep_lines <- sort(unique(keep_lines))
if (all(keep_lines > 0)) {
keep_string <- citfile_clean[keep_lines]
citfile_clean[keep_lines] <- ""
}
# Remove rest of brackets
citfile_clean <- gsub('["', '"', citfile_clean, fixed = TRUE)
citfile_clean <- gsub('"]', '"', citfile_clean, fixed = TRUE)
if (all(keep_lines > 0)) {
# Add "good" brackets back
citfile_clean[keep_lines] <- keep_string
}
writeLines(citfile_clean, cit_temp)
# Download latest scheme
schema_temp <- tempfile("schema", fileext = ".json")
download.file("https://raw.githubusercontent.com/citation-file-format/citation-file-format/main/schema.json",
schema_temp,
mode = "wb", quiet = TRUE
)
# Validate
result <- jsonvalidate::json_validate(cit_temp,
schema_temp,
verbose = TRUE
)
# Results
message("------\n")
#> ------
if (result == FALSE) {
print(knitr::kable(attributes(result)$errors,
align = "l",
caption = paste(citation_path, "errors:")
))
message("\n\n------")
stop(citation_path, "file not valid. See Artifact: citation-cff-errors for details.")
} else {
message(citation_path, "is valid.")
message("\n\n------")
}
#>
#>
#> Table: ./key-error/CITATION.cff errors:
#>
#> |field |message |
#> |:---------------|:--------------------------------|
#> |data |has additional properties |
#> |data.authors.0 |no schemas match |
#> |data.doi |referenced schema does not match |
#> |data.keywords.0 |is the wrong type |
#> |data.license |referenced schema does not match |
#> |data.url |referenced schema does not match |
#>
#>
#> ------
#> Error in eval(expr, envir, enclos): ./key-error/CITATION.cfffile not valid. See Artifact: citation-cff-errors for details.Created on 2021-09-06 by the reprex package (v2.0.1)
References
Druskat, S., Spaaks, J. H., Chue Hong, N., Haines, R., Baker, J., Bliven, S., Willighagen, E., Pérez-Suárez, D., & Konovalov, A. (2021). Citation File Format (Version 1.2.0) [Computer software]. https://doi.org/10.5281/zenodo.5171937