This documentation describes v0.2.0 of
- the base DeepPhe system, which
- extracts information from plaintext documents using Apache cTAKES™
- summarizes information for Cancers and Tumors across mutiple documents
- writes results to a database
- and DeepPhe Viz, for visualizing the cancer patient summaries generated by the DeepPhe system.
The output of DeepPhe has elements that are specific to the following three cancer domains:
- Breast Cancer
- Ovarian Cancer
- Malignant Melanoma
The system has been tested against documents from those three domains.
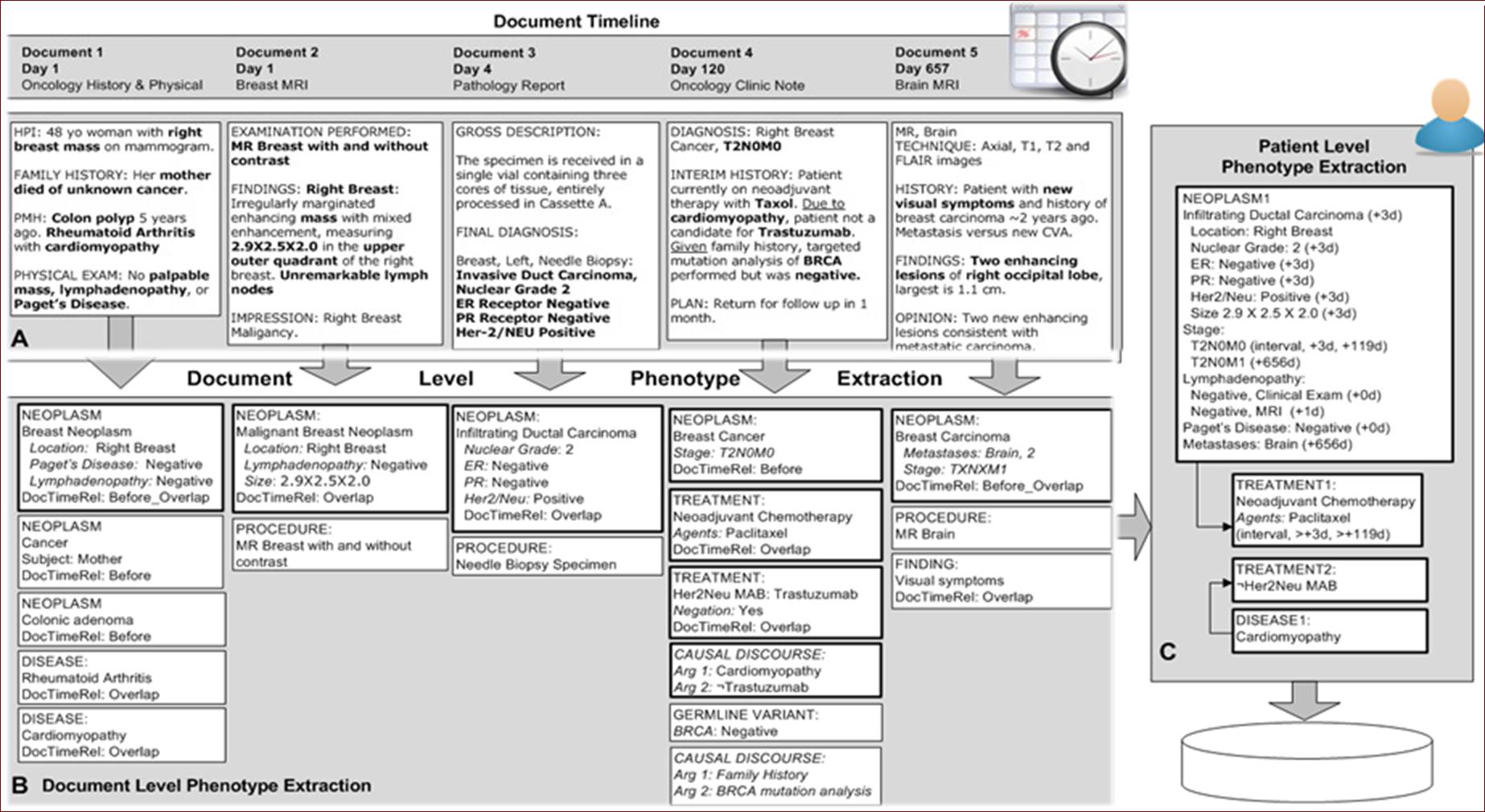
Here is a pictorial example of DeepPhe processing five documents for a single patient, and summarizing the cancer information from the five documents. Some of the attributes, such as the tumor size and treatment, show future direction of DeepPhe beyond version 0.2.0.
- Install the base DeepPhe system.
- Install DeepPhe-Viz, the DeepPhe Visualizer.
- Preparing the documents you would like to process
- Save the documents as plaintext files, one directory per patient
- Naming the files you would like to process
- Configuring your input and output directories
- Telling the system which cancer domain your files belong to
- Listing the section headings used by your institution
- Running the DeepPhe system
- Inspecting the output files
- Viewing the results using DeepPhe-Viz
- Accessing the neo4j database directly
- This release of DeepPhe uses neo4j 3.2.x. WARNING - do not simply download the latest version of neo4j.
Visit the DeepPhe wiki for more.
DeepPhe is provided under an Academic Software Use Agreement
Refer to that agreement for information about requesting the use of the Software for commercial purposes.
DeepPhe includes portions of the HemOnc.org ontology. Refer to HemOnc.org regarding the licensing of the HemOnc.org ontology.
DeepPhe includes portions of the NCI Thesaurus (NCIt).
The viz plugin (refer to deepphe-viz-neo4j) has a dependency on apoc.
Other licenses for your reference
- Apache cTAKES™
- jchronic
- Neo4j
- Drools
Please drop us a note if you obtain the code, by posting to the DeepPhe group.
Metrics on downloads and usage could help us with funding future enhancements.
For questions, contact us via the DeepPhe group.