MendelianRNA-seq-DB was developed to help researchers discover abnormal transcripts causitive for neuromuscular disease. It is a tool which analyzes junction positions in a collection of BAM files.
The scripts are a rewrite of those used in the study: Improving genetic diagnosis in Mendelian disease with transcriptome sequencing by Cummings et. al. (2017) to support parallel processing and storing junction information in a database. The use of parallel processing can lower runtime by a factor of 6 when using a Sandy Bridge 10 core Intel machine. The use of a junction position database confers a few benefits:
- BAM files only have to be processed once through SpliceJunctionDiscovery, and results from previous computations can be reused
- The ability to utilize a very high number of controls, possibly up to the thousands
- Lowered RAM use since junction positions are being stored on disk
- Generate 2 sets of splice junction positions from a collection of .bam files. One set is considered to be "healthy" and the other is considered to be "disease"
- Remove any shared splice junction positions from the "disease" set since splicing events causitive for disease are likely not present in a "healthy" population (keep in mind we are dealing with muscular dystrophy - a rare disease)
- Remove splice sites from the "disease" set which have a low number of read counts and/or normalized read counts and thus can considered as noise
- Priortize and analyze remaining junctions which reside in genes related to this disease
In my experience, it is possible to narrow down the number of candidate splice sites from ~240 000 to 30 or less per sample using these scripts.
It follows that with a higher number of control BAM files you are able to filter out more non-pathogenic and noise junctions leading to a smaller number of candidate splicing events. This is clearly seen in a graph created by Beryl Cummings:
Ideally, you want to use as many controls as you can. In practice, you may want to rely on a few tricks to reduce your dataset to a size where you can actually analyze each sample specific site on IGV:
- Don't analyze junctions annotated with 'BOTH'. This is the "safest" trick and one that you should probably always use. 'BOTH' annotated junctions are junctions which are seen in your transcript_model files (gencode v19).
- Construct a gene panel, from experts or from scientific literature, and only analyze junctions pertaining to those regions
- Set a higher threshold for read counts. This can be specified as a parameter when running FilterSpliceJunctions.py
- If you believe you have a high enough read depth across regions of interest in the transcriptome don't analyze junctions annotated with 'NONE'. This is the most "dangerous" trick. It removes all possibility of discovering splicing events which start and end in a known exonic regions and a few other edge cases.
SpliceJunctionDiscovery.py calls upon samtools to report the presence of introns in a list of regions of interest, determines their chromosomal positions, counts the number of alignments to each position, and writes this to a text file (e.x. PATIENT_X/DMD.txt).
AddJunctionsToDatabase.py reads each text file, compares the reported junctions to that of a transcript_model, annotates any shared splice site positions, normalizes annotated reads, and stores this information into a database.
FilterSpliceJunctions.py contains some pre-defined queries which can be used to filter junctions in the database in hopes of finding an aberrant splicing event causative for disease.
SpliceJunctionDiscovery.py usually takes the longest to execute. This step is parallelized and the number of worker processes can specified in the torque file or as an arguement to the script. AddJunctionsToDatabase.py is much faster and likely takes minutes to an hour for sample sizes less than 100. Querrying the database using FilterSpliceJunctions is probably the fastest step taking seconds to execute.
-
.bam (and .bai) files produced from an RNA-seq pipeline - All control or "healthy" .bams need to have the phrase 'GTEX' in their file name for read count logic to work properly. The GTEx project is what I used for control BAMs. All BAM files in the database should be from the same tissue due to tissue specific expression.
-
transcript_file - A tab-delimited text file containing a list of genes and their spanning chromosome positions that you want to discover junctions in. The format of the file is:
GENE ENSG STRAND CHROM START STOP GENE_TYPEYou can use genes.R for that, or convert an existing .bed file using this bash line:
cat all-protein-coding-genes.bed | awk '{print $4"\t"$4"\t+\t"$1"\t"$2"\t"$3"\tNEXONS"}' >> all-protein-coding-genes.listThere is an included file which contains all protein coding regions.
-
bamlist.list - A file containing the names of all the bams you want to discover junctions in. The file should quite simply be:
G65693.GTEX.8TY6-5R5T.2.bam G55612.GTEX.7G54-3GS8.1.bam G09321.GTEX.0EYJ-9E12.3.bam PATIENT.bamAn easy way to generate this file would be to navigate to a directory containing the .bam files you want to use and running this line:
ls *.bam | grep '' > bamlist.list -
transcript_model - A text file containing a list of known canonical splice junctions. These will be used to evaluate a junction's annotation (NONE, START, STOP, BOTH, EXON_SKIP). You can use your own, or use the included file. This file contains junctions from gencode v19.
-
Python 3.5.2 or higher
-
Python CIGAR string library by Brent Pedersen
-
sqlite3 Python library based off of SQLite3 version 3.11.0 or higher. You can check your library's version with:
import sqlite3 print (sqlite3.sqlite_version_info)
-
Put bamlist.list, .bam files, .bai files in a new directory. Navigate to it.
-
For Torque users there is a PBS file. Just change the "home" directory in the file to match where you placed the MendelianRNA-seq-DB folder and run:
qsub MendelianRNA-seq/Analysis/rnaseq.novel_splice_junction_discovery.pbs -v transcript_file=transcript_file,bam_list=bamlist.list,processes=10For non-Torque users, SpliceJunctionDiscovery can be run from terminal:
python3 MendelianRNA-seq/Analysis/SpliceJunctionDiscovery.py -transcript_file=$transcript_file -bam_list=$bam_list -processes=$processesParameters:
-
transcript_file, path to file #2
-
bam_list, path to file #3
-
processes, the number of worker processes running in the background calling samtools. This number should be equal to or less than the number of cores on your machine.
For torque users: This number should also be equal to or less than the number specified for ppn in rnaseq.novel_splice_junction_discovery.pbs:
#PBS -l walltime=10:00:00,nodes=1:ppn=10
-
-
Run AddJunctionsToDatabase.py with --addGencode to initally populate the database with gencode junctions.
python3 AddJunctionsToDatabase.py --addGencode -transcript_model=gencode.comprehensive.splice.junctions.txt -
Run AddJunctionsToDatabase.py with --addBAM to populate the database with junctions and read counts from your samples.
python3 AddJunctionsToDatabase.py --addBAM -transcript_file=all-protein-coding-genes-no-patches.txt -processes=4 -bamlist=bamlist.list -flank=1-flank is a parameter which specifies a flanking region for transcript_model annotation. If flank was set to 1, a gencode junction was 1:100-300 and a junction in a sample was 1:99-301, the sample junction would be considered BOTH annotated. This is because both the start and stop positions fall within a +/- 1 range of that of a transcript_model's junction.
-
Now you can use FilterSpliceJunction.py to output junction information.
To print out splice sites only seen in a "disease" sample and not in any GTEx sample use:
python3 FilterSpliceJunctions.py --sample [SAMPLE_NAME] [MIN_READ_COUNT] [MIN_NORM_READ_COUNT]I typically use the following values:
[MIN_READ_COUNT] = 5 [MIN_NORM_READ_COUNT] = 0.05Note: Because the query in the
--sampleoption joins information from a single sample's name, columnssample:read_countandsample:norm_read_countwill not show read counts from other samples. This is not the case with the---alloption however.To print out splice sites across all samples in the database, use:
python3 FilterSpliceJunctions.py --allYou may want to use awk and grep tools on the
--alltext file to perform more complex filters and to avoid writing your own database queries.Further documentation on how to interpret these results can be found in a blog post written by Beryl Cummings.
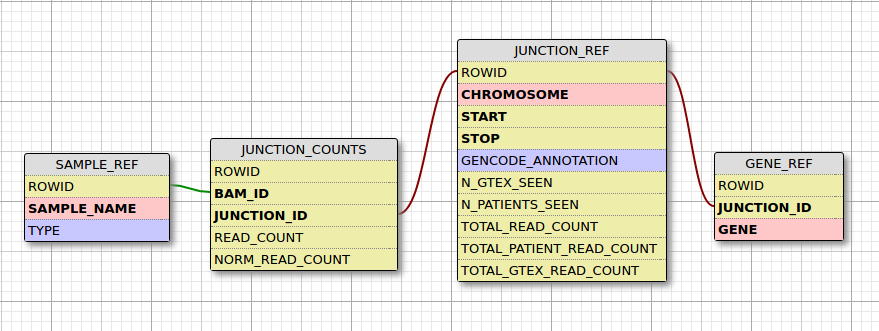
By default the database is named SpliceJunction.db. There are 4 tables:
1. SAMPLE_REF, a list of samples and their type (0 = GTEX or control, 1 = patient)
2. JUNCTION_REF, a list of junctions and their frequency of appearances in samples
3. JUNCTION_COUNTS, read counts of junctions in a sample
4. GENE_REF, an annotation of junctions with genes, a single junction can map to multiple genes
Using one of the options of FilterSpliceJunctions.py will produce a text file containing junction information in the following format:
gene chromosome:start-stop annotation n_gtex_seen n_patients_seen total_patient_read_count total_gtex_read_count total_read_count sample:read_count sample:norm_read_count
MT-ND1 MT:3540-3610 NONE 0 1 11 0 11 PATIENT.bam:11 PATIENT.bam:NULL
AC002321.2 GL000201.1:4130-9415 NONE 1 1 32 4 36 PATIENT.bam:32 PATIENT.bam:NULL
MT-CO1 MT:7276-13822 NONE 1 1 5 1 6 PATIENT.bam:5 PATIENT.bam:NULL
MT-ATP6 MT:9234-9511 NONE 0 1 6 0 6 PATIENT.bam:6 PATIENT.bam:NULL
AC002321.2 GL000201.1:9511-14322 START 1 1 70 2 72 PATIENT.bam:70 PATIENT.bam:NULL
- SpliceJunctionDiscovery has been rewritten in Python and parallelized - decreasing processing time by a factor proprotional to the number of worker processes
- CIGAR string parsing is handled by a function called parseCIGARForIntrons() whereas before CIGAR strings were handled by piping through multiple bash tools. As a result of improper parsing using bash tools, junction start and/or stop positions were not reported properly (e.x. 1:100-200*1D30 represents an alignment that should really be 1:100-230 or 1:100-231)
- Transcript_model annotation and flanking have been implemented using database logic
- All information produced by SpliceJunctionDiscovery is stored in a database instead of text files
- The database has some new fields that can be used to filter junctions:
n_patients_seen n_gtex_seen total_read_count total_patient_read_count total_gtex_read_count
- Transcript_model annotation now discriminates between 'START' and 'STOP' instead of 'ONE'. In addition, there is a new annotation, called 'EXON_SKIP' which denotes the event of exon skipping. This is done by checking to see if the reported 3' and 5' positions from a sample's junction belong to different transcript_model junctions
- Normalization of annotated junctions now considers read counts from all junctions that have at least one annotated splice site as the denominator whereas before only "BOTH" annotated junctions were used
Improving genetic diagnosis in Mendelian disease with transcriptome sequencing
RNAseq analysis for the diagnosis of muscular dystrophy
The original scripts: MendelianRNA-seq
Blog post on how to interpret reported junction positions
A single gene region as defined in the format (CHROMOSOME:START-STOP) can encompass partial or whole regions of other genes. Thus, the same junction can appear in 2 different gene text files in a sample folder generated by SpliceJunctionDiscovery. Whether a junction belongs to two or more genes is not always factually correct. However, for the sake of inclusion and for the fact that this rarely happens, this edge case has been accounted for in AddJunctionsToDatabase.py in two ways:
1. The mapping of a single junction to multiple genes has been done with the table GENE_REF
2. If the script encounters the same junction in a sample more than once, it will utilize the result with the highest read count for read count and normalized read count and will discard the other.
A +/- flanking region is considered when annotating the 5' and 3' positions of sample junctions to increase the number of annotated junctions. This value is specified by the -flank parameter (default 1). There is an option to not use flanking at all (-flank 0).
In order to circumvent the issue of write locks each worker process in SpliceJunctionDiscovery is assigned a single gene and writes to a single text file. As a result, each sample folder contains around 15000 to 22000 gene text files if you were to run the pipeline against all protein coding genes.
Using a DFS does not affect the performance of SpliceJunctionDiscovery, however, it does affect AddJunctionsToDatabase significantly. Because the script opens, reads, and closes many small files, using a DFS will result in a majority of runtime spent looking for these files on the server. In my experience, this increased runtime from 5 minutes (on a local SSD) to over 40 hours (on the server). Therefore, it is reccomended that you copy over the files created by SpliceJunctionDiscovery to a local drive or simply generate them on a local drive before running AddJunctionsToDatabase.