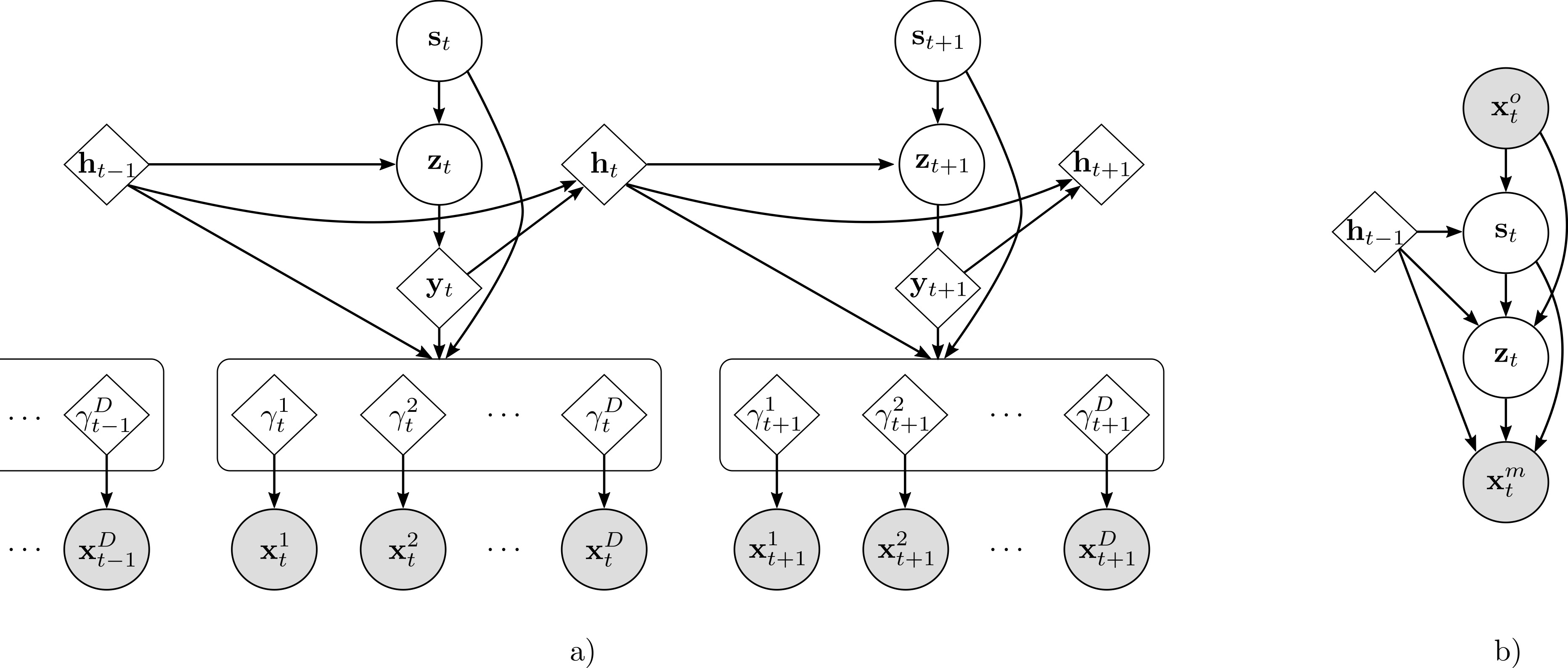
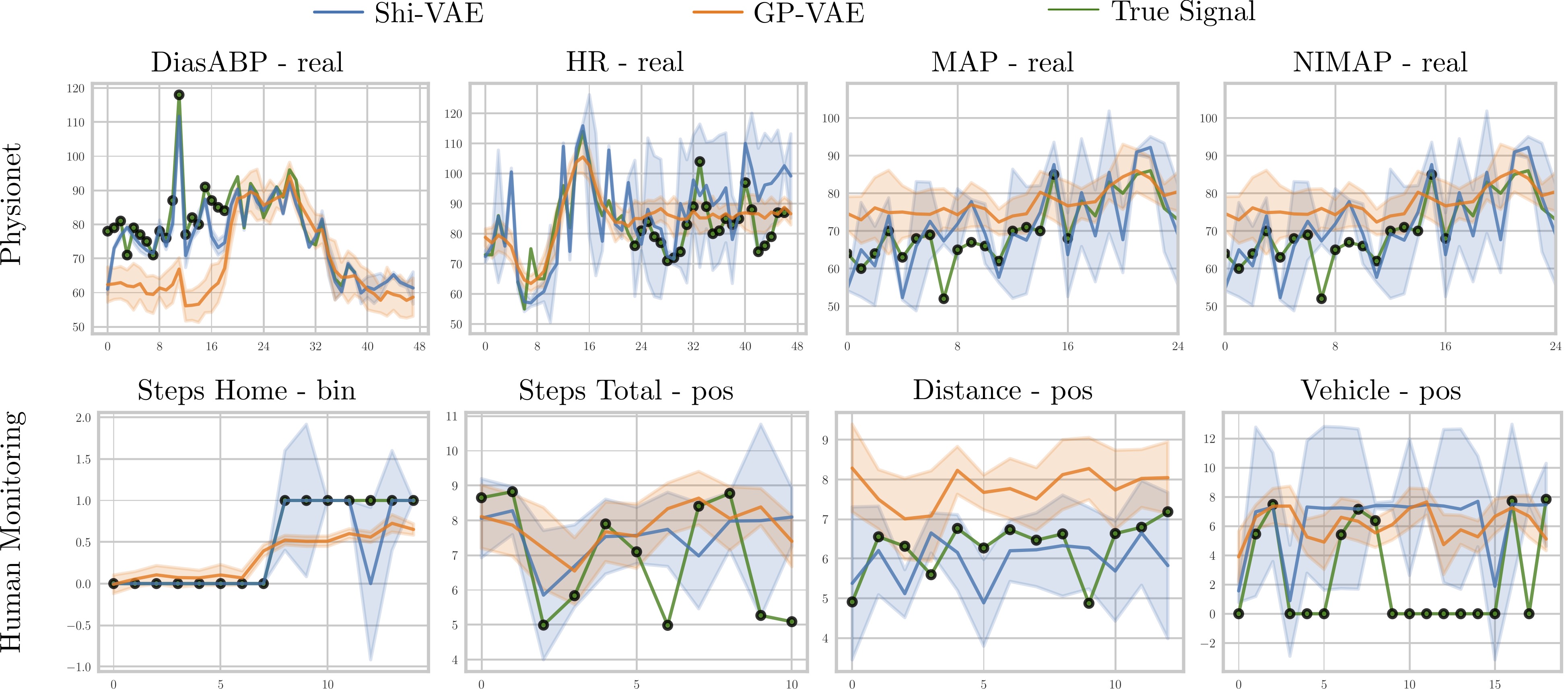
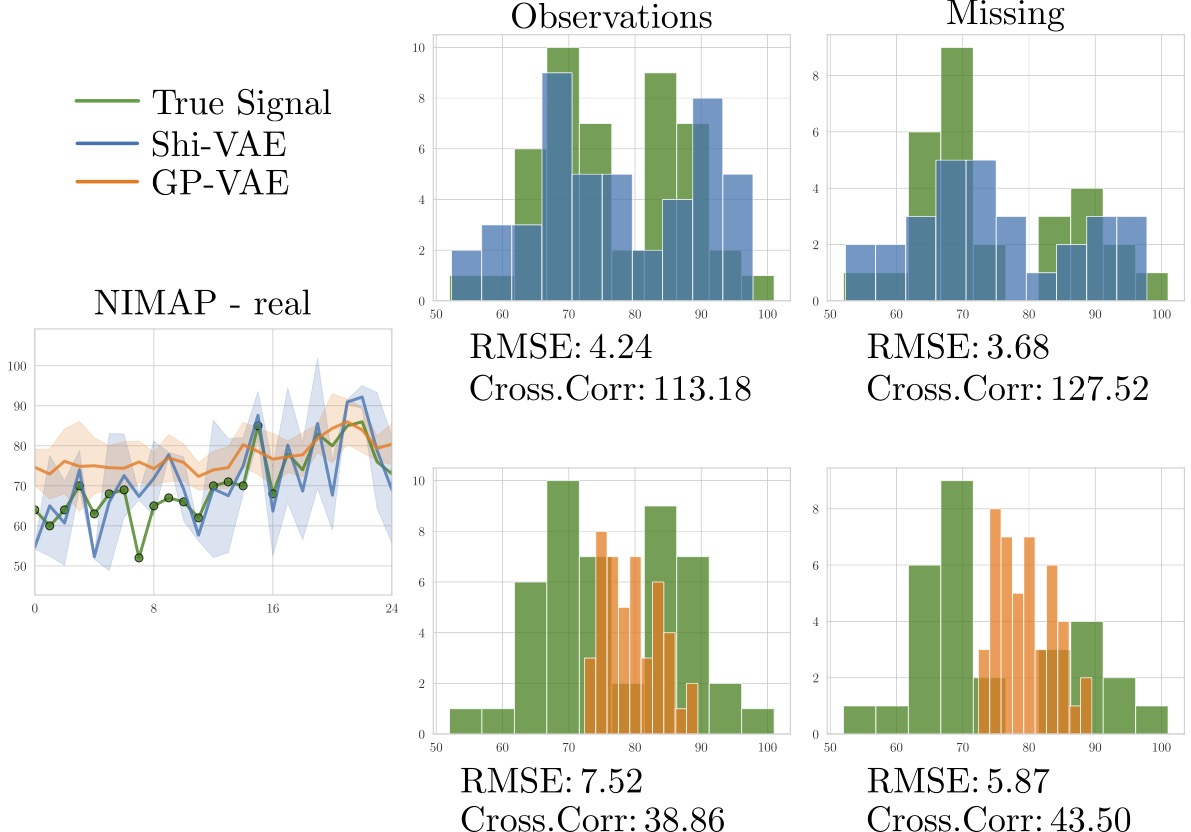
Code implementation for the paper Medical data wrangling with sequential variational autoencoders. Preprint also available in arxiv. Please, if you use this code, cite the paper using:
@article{shivae,
author={Barrejon, Daniel and M. Olmos, Pablo and Artes-Rodriguez, Antonio},
journal={IEEE Journal of Biomedical and Health Informatics},
title={Medical data wrangling with sequential variational autoencoders},
year={2021},
volume={},
number={},
pages={1-1},
doi={10.1109/JBHI.2021.3123839}}
The installation can be done using Conda:
conda env create -f environment.yml
conda activate shivae
or using the requirements.txt. Using pip, it can be installed with the command below:
conda create -n shivae python=3.7
conda activate shivae
pip install -r requirements.txt
In data/, you can find an already created synthetic dataset using an Heterogeneous HMM.
If you want to create a custom heterogeneous HMM dataset, go to the README.md document inside
hmm_dataset.
In data/physionet_burst/ unzip the zip file:
tar -xvf physionet_burst.zip
This zip file contains 4 files:
-
data_types.csv: CSV file containing the information for all the attributes. -
data_types_real.csv: CSV file containing the information for all the attributes, assuming only real values. -
data_types_pos.csv: CSV file containing the information for all the attributes, assuming only positive values. -
physionet_burst.npz: NPZ file containing:x_{set}_full: Data for {set} containing all the observed attributes.x_{set}_miss: Data for {set} containing observed values, except those values used as artificial missing for evaluation.m_{set}_miss: Missing mask for {set}, containing real missing and artificial missing.x_{set}_artificial: Mask for {set} at those observed values used for evaluation.y_{set}: y value for {set}.
For running all the experiments, always use the src folder as working directory.
Some examples are included in main_hmm.py and main_physionet.py.
You can run the following command to run an example on the synthetic dataset:
python3 main_hmm.py --experiment hmm --train -1 --n_epochs 100 --z_dim 2 --K 3 --kl_annealing_epochs 20 --percent_miss 30
In case you want to check the performance with the physionet dataset, run this example:
python3 main_physionet.py --experiment physionet --train -1 --n_epochs 100 --z_dim 35 --K 10 --kl_annealing_epochs 20
By running these examples you will train the models and check the performance with some images generated for
reconstruction and generation.
For running the scripts used in the paper, you can just run
# Physionet Results, inside /src:
python3 results_hmm.py --train -1
# Physionet Results, inside /src:
python3 results_physionet.py --train -1
Using the --help option displays the following information.
usage: main_hmm.py [-h] [--model MODEL] [--train TRAIN] [--restore RESTORE]
[--experiment EXPERIMENT] [--n_epochs N_EPOCHS]
[--kl_annealing_epochs KL_ANNEALING_EPOCHS]
[--l_rate L_RATE] [--percent_miss PERCENT_MISS]
[--save_model SAVE_MODEL] [--save_every SAVE_EVERY]
[--print_every PRINT_EVERY] [--plot_every PLOT_EVERY]
[--learn_std LEARN_STD] [--z_dim Z_DIM] [--h_dim H_DIM]
[--K K] [--local LOCAL] [--gpu GPU]
[--batch_size BATCH_SIZE] [--result_imgs RESULT_IMGS]
[--clip CLIP] [--data_dir DATA_DIR]
[--filter_vars FILTER_VARS [FILTER_VARS ...]]
[--dataset DATASET] [--ckpt_dir CKPT_DIR]
[--ckpt_file CKPT_FILE]
optional arguments:
-h, --help show this help message and exit
--model MODEL Used model.
--train TRAIN Training options [0: Results, 1:Train, -1:Train and
Result]
--restore RESTORE Restore training
--experiment EXPERIMENT
Experiment name.
--n_epochs N_EPOCHS Number of epochs to train
--kl_annealing_epochs KL_ANNEALING_EPOCHS
Number of epochs to apply annealing on KL divergence.
--l_rate L_RATE Learning rate
--percent_miss PERCENT_MISS
Missing rate for HMM dataset, or any synthetic
dataset.
--save_model SAVE_MODEL
Save model.
--save_every SAVE_EVERY
Save model every n epochs
--print_every PRINT_EVERY
Print model every n epochs
--plot_every PLOT_EVERY
Plot ELBO every n epochs
--learn_std LEARN_STD
Learn std for real and positive distributions.
--z_dim Z_DIM Dimension of z
--h_dim H_DIM Dimension of h
--K K Dimension of latent code s for ShiVAE
--local LOCAL Set to 1 for local execution. 0 for cluster/server
execution.
--gpu GPU Select gpu=-1: CPU. Else: GPU.
--batch_size BATCH_SIZE
Number of samples per batch-
--result_imgs RESULT_IMGS
Number of images to show in reconstruction.
--clip CLIP Clipping value to avoid exploding gradients.
--data_dir DATA_DIR Parent directory for data.
--filter_vars FILTER_VARS [FILTER_VARS ...]
Variables to use. Use None for using all variabes.
--dataset DATASET Name of the dataset for the experiment.
--ckpt_dir CKPT_DIR Parent directory to save models
--ckpt_file CKPT_FILE
Pytorch Weights (.pth), with absolute path.
Daniel Barrejon
Pablo M. Olmos
Antonio Artés-Rodríguez
For any question regarding the code or the model, please send an email to: dbarrejo@ing.uc3m.es