Nipoppy is a lightweight framework for standardized organization and processing of neuroimaging-clinical datasets. Its goal is to help users adopt the FAIR principles and improve the reproducibility of studies.
The framework includes three components:
-
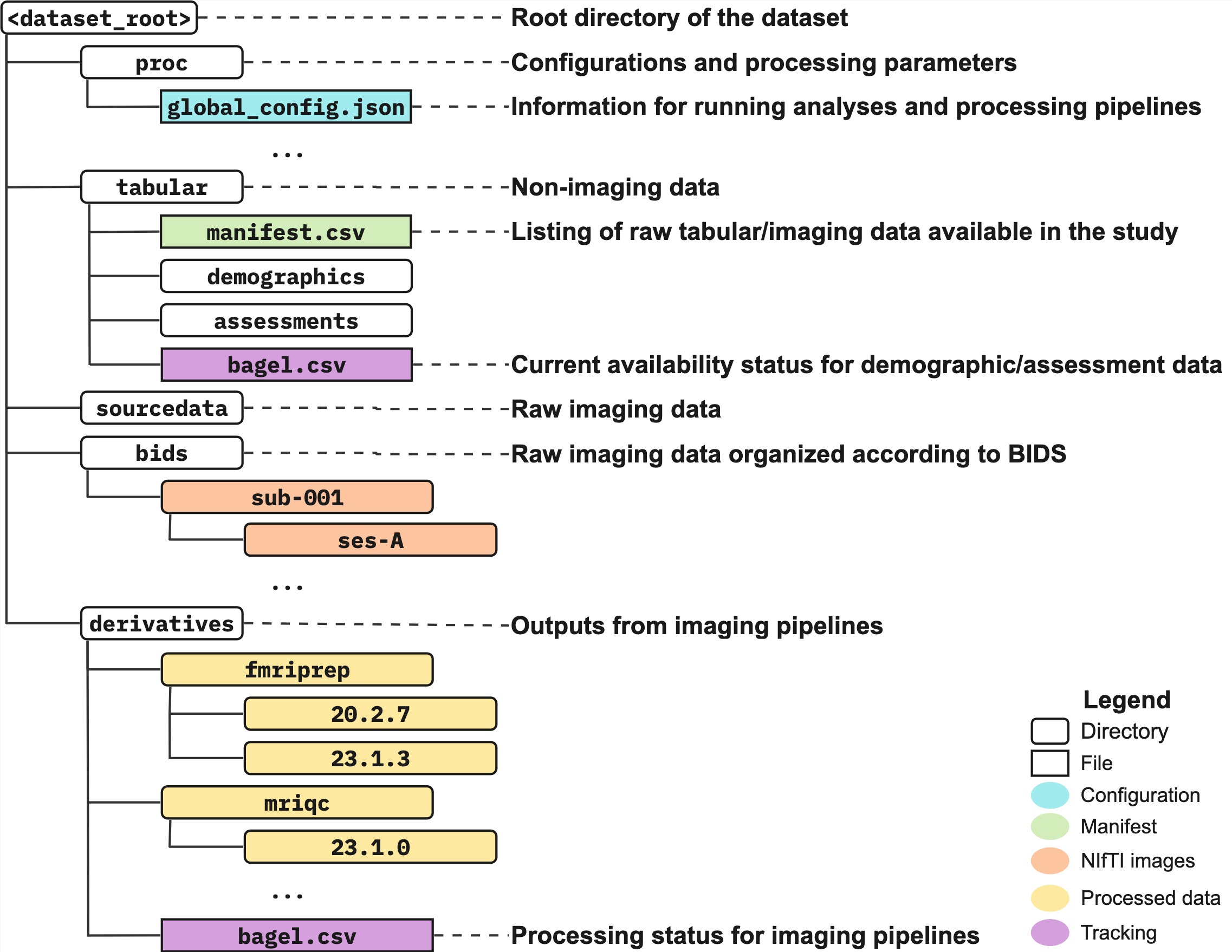
A specification for dataset organization that extends the Brain Imaging Data Structure (BIDS) standard by providing additional guidelines for tabular (e.g., phenotypic) data and imaging derivatives.
-
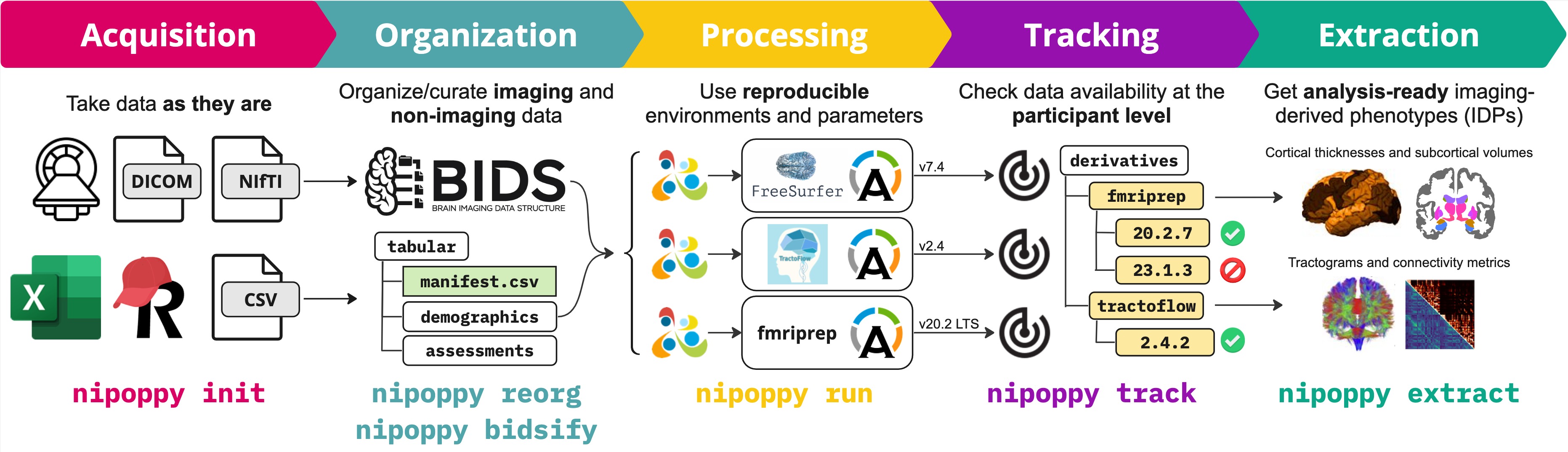
A protocol for data organization, curation and processing, with steps that include the following:
- Organization of raw data, including conversion of raw DICOMs (or NIfTIs) to BIDS
- Processing of imaging data with existing or custom pipelines
- Tracking of data availability and processing status
- Extraction of imaging-derived phenotypes (IDPs) for downstream statistical modelling and analysis
-
A command-line interface and Python package that provide user-friendly tools for applying the framework. The tools build upon existing technologies such as the Apptainer container platform and the Boutiques descriptor framework. Several existing containerized pipelines are supported out-of-the-box, and new pipelines can be added easily by the user.
- We have also developed a web dashboard for interactive visualizations of imaging and phenotypic data availability.
See the documentation website for more information!