README
Nextflow enables scalable and reproducible scientific workflows using software containers.
Installation
Conda is an easy way of installing Nextflow. I would recommend using Mamba, which is a faster version of Conda, to install Nextflow.
mamba create -y \
-n nextflow \
-c Bioconda -c conda-forge \
nextflow
conda activate nextflow
nextflow -version
#
# N E X T F L O W
# version 22.10.6 build 5843
# created 23-01-2023 23:20 UTC (24-01-2023 08:20 JDT)
# cite doi:10.1038/nbt.3820
# http://nextflow.io
#
java -version
# openjdk version "17.0.3-internal" 2022-04-19
# OpenJDK Runtime Environment (build 17.0.3-internal+0-adhoc..src)
# OpenJDK 64-Bit Server VM (build 17.0.3-internal+0-adhoc..src, mixed mode, sharing)Set up for testing purposes only
-
Java 11 or later is required (up to 19 is supported);
script/install_openjdk.shwill install OpenJDK 19 intobin. Do not install version 20 or you will get the following error:ERROR: Cannot find Java or it's a wrong version -- please make sure that Java 8 or later (up to 19) is installed
script/install_openjdk.sh
bin/jdk-19.0.2/bin/java -version
# openjdk version "19.0.2" 2023-01-17
# OpenJDK Runtime Environment (build 19.0.2+7-44)
# OpenJDK 64-Bit Server VM (build 19.0.2+7-44, mixed mode, sharing)- Install Nextflow.
export PATH=$(pwd)/bin/jdk-19.0.2/bin:$PATH
curl -s https://get.nextflow.io | bash
# snipped
# N E X T F L O W
# version 23.04.0 build 5857
# created 01-04-2023 21:09 UTC (02-04-2023 06:09 JDT)
# cite doi:10.1038/nbt.3820
# http://nextflow.io
#
# Nextflow installation completed. Please note:
# - the executable file `nextflow` has been created in the folder: /home/dtang/github/learning_nextflow
# - you may complete the installation by moving it to a directory in your $PATH
mv nextflow bin/jdk-19.0.2/bin/- Hello world example.
nextflow run hello
# N E X T F L O W ~ version 23.04.0
# Pulling nextflow-io/hello ...
# downloaded from https://github.com/nextflow-io/hello.git
# Launching `https://github.com/nextflow-io/hello` [fervent_mendel] DSL2 - revision: 1d71f857bb [master]
# executor > local (4)
# [ae/272202] process > sayHello (4) [100%] 4 of 4 ✔
# Bonjour world!
#
# Ciao world!
#
# Hello world!
#
# Hola world!Nextflow tutorial
Introduction to Bioinformatics workflows with Nextflow and nf-core lesson objectives:
- The learner will understand the fundamental components of a Nextflow script, including channels, processes and operators.
- The learner will write a multi-step workflow script to align, quantify, and perform QC on an RNA-Seq data in Nextflow DSL.
- The learner will be able to write a Nextflow configuration file to alter the computational resources allocated to a process.
- The learner will use nf-core to run a community curated pipeline, on an RNA-Seq dataset.
Nextflow is a workflow management system that combines a runtime environment, software that is designed to run other software, and a programming domain specific language (DSL) that eases the writing of computational pipelines.
Nextflow is built around the idea that Linux is the lingua franca of data science. Nextflow follows Linux's "small pieces loosely joined" philosophy, in which many simple but powerful command-line and scripting tools, when chained together, facilitate more complex data manipulations.
Nextflow extends this approach, adding the ability to define complex program
interactions and an accessible (high-level) parallel computational environment
based on the dataflow programming model, whereby processes are connected via
their outputs and inputs to other processes, and run as soon as they
receive an input.
The Nextflow scripting language is an extension of the Groovy programming language, which in turn is a superset of the Java programming language. Groovy simplifies the writing of code and is more approachable than Java; see Groovy's semantics.
Nextflow (version > 20.07.1) provides a revised syntax to the original DSL, known as DSL2. This feature is enabled by the following directive at the beginning of a workflow script:
nextflow.enable.dsl=2
Processes, channels, and workflows
Nextflow workflows have three main parts:
- Processes - describe a task to be run and can be written in any scripting language that can be executed on Linux (e.g. Bash, Perl, Python, etc.). They define inputs and outputs for a task. Processes spawn a task for each complete input set and each task is executed independently and cannot interact with another task. The only way data can be passed between process tasks is via asynchronous queues, called channels.
- Channels - channels are used to manipulate the flow of data from one process to the next.
- Workflows - this section is used to explicitly define the interaction between processes and the pipeline execution flow.
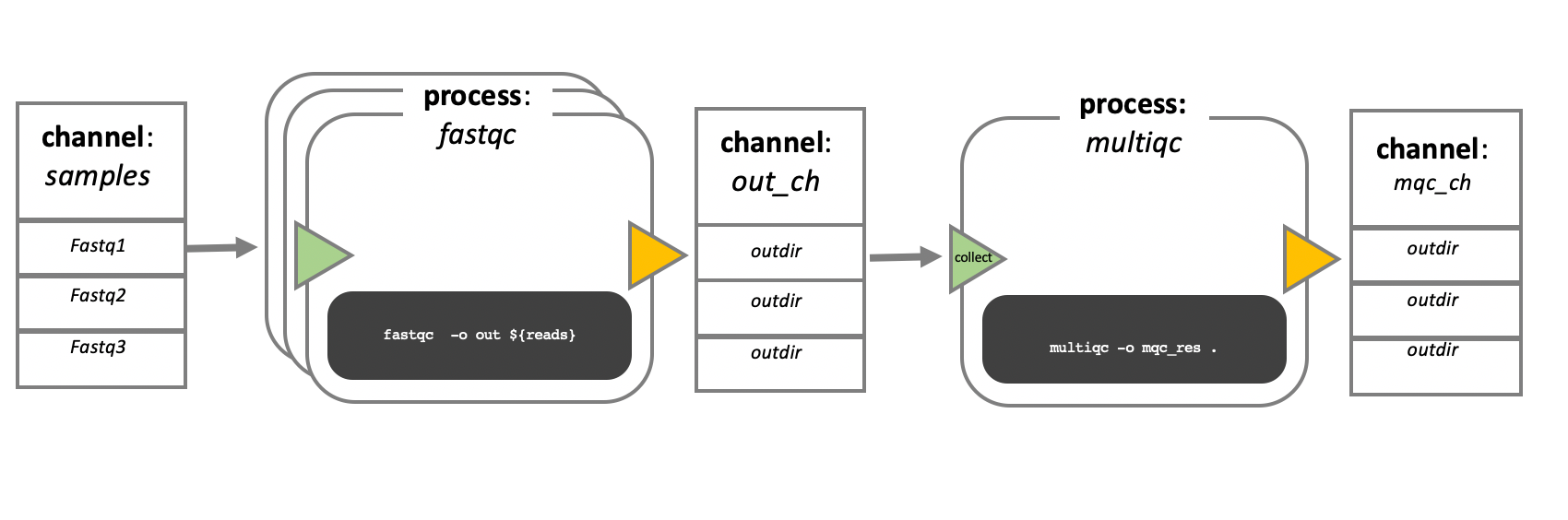
In the example above we have a samples channel containing three input FASTQ
files. The fastqc process takes the samples channel as input and since the
channel has three elements, three independent instances (tasks) of that process
are run in parallel. Each task generates an output, which is passed to the
out_ch channel and is used as input for the multiqc process.
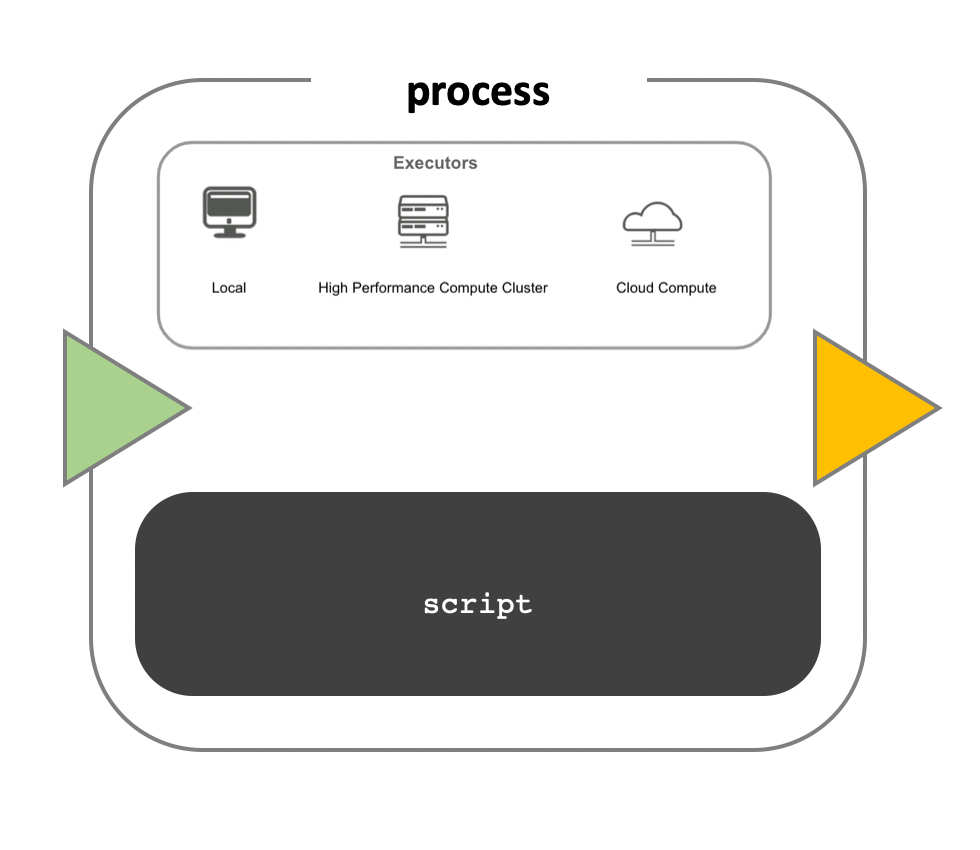
While a process defines what command or script has to be executed, the
executor determines how that script will be run in the target system. The
default is to execute processes on the local computer, which is useful for
pipeline development, testing, and small scale workflows. For large scale
computational pipelines, a High Performance Cluster (HPC) or Cloud platform is
desired.
Nextflow provides a separation between the pipeline's functional logic and the underlying execution platform. This makes it easy to execute a pipeline on various platforms without modifying the workflow and is achieved by defining the target execution platform in a configuration file.
Nextflow provides native support for major batch schedulers and cloud platforms including SGE, SLURM, AWS, and Kubernetes.
First script
The wc.nf script counts the number of lines in a FASTQ file.
nextflow run script/wc.nf
# N E X T F L O W ~ version 23.04.0
# Launching `script/wc.nf` [stoic_easley] DSL2 - revision: 9cef523068
# executor > local (1)
# [6b/286e0b] process > NUM_LINES (1) [100%] 1 of 1 ✔
# ref1_1.fq.gz 58708Sarek
nf-core/sarek is a Nextflow workflow for calling variants on whole genome, exome, or targeted sequencing data. Following the Quick Start guide:
- Install using Mamba.
- Docker installed.
- Download and test pipeline:
time nextflow run nf-core/sarek -profile test,docker --outdir sarek_test
# snipped
# -[nf-core/sarek] Pipeline completed successfully-
# Completed at: 14-Apr-2023 09:48:56
# Duration : 9m 18s
# CPU hours : 0.1
# Succeeded : 26
#
# real 9m37.830s
# user 1m51.006s
# sys 0m14.591s- Run your own analysis
nextflow run nf-core/sarek --input samplesheet.csv --outdir <OUTDIR> --genome GATK.GRCh38 -profile dockerSee the usage page for more information.