Iva Filipovic1,2,3, Laura Chiossone4, Paola Vacca5, Russell S. Hamilton2,3, Jean-Marc Doisne1,6, Gerard Eberl6, Thierry Walzer7, Cristina Mingari5, Andrew Sharkey3,8, Lorenzo Moretta9 & Francesco Colucci1,3,§
1 Department of Obstetrics and Gynaecology, University of Cambridge, University of Cambridge School of Clinical Medicine, NIHR Cambridge Biomedical Research Centre, UK
2 Department of Physiology, Development and Neuroscience, University of Cambridge, UK
3 Centre for Trophoblast Research, University of Cambridge, UK
4 G. Gaslini Institute, Genoa, Italy
5 Department of Experimental Medicine (DIMES), University of Genoa, Italy
6 Department of Immunology, Pasteur Institute, Paris, France
7 Centre International de Recherche en Infectiologie, INSERM U1111, Lyon, France
8 Department of Pathology, University of Cambridge, UK
9 Department of Immunology, IRCCS Bambino Gesù Children’s Hospital, Rome, Italy
§ Corresponding author: fc287@medschl.cam.ac.uk
Filipovic, I, Chiossone, L., Vacca, P., Hamilton, R.S., Doisne, J.M., Eberl, G., Walzer, T., Mingari, C., Sharkey, A., Moretta, L., Colucci, F. (2018) Molecular definition of group 1 innate lymphoid cells in the mouse uterus. [Nature Communications, 9, 4492] [DOI]
Determining the function of uterine lymphocytes is challenging because of the rapidly changing nature of the organ in response to sex hormones and, during pregnancy, to the invading fetal trophoblast cells. Here we provide the first genome-wide transcriptome atlas of mouse uterine group 1 innate lymphoid cells (g1 ILCs) at mid-gestation. The composition of g1 ILCs fluctuates throughout reproductive life, with Eomes-veCD49a+ ILC1s dominating before puberty and specifically expanding in second pregnancies, when the expression of CXCR6, a marker of memory cells, is upregulated. Tissue-resident Eomes+CD49a+ NK cells (trNK), which resemble human uterine NK cells, are most abundant during early pregnancy, and showcase gene signatures of responsiveness to TGF-β, connections with trophoblast, epithelial, endothelial and smooth muscle cells, leucocytes, as well as extracellular matrix. Unexpectedly, trNK cells express genes involved in anaerobic glycolysis, lipid metabolism, iron transport, protein ubiquitination, and recognition of microbial molecular patterns. Conventional NK cells expand late in gestation and may engage in crosstalk with trNK cells involving IL-18 and IFN-γ. These results identify trNK cells as the cellular hub of uterine g1 ILCs at mid-gestation and mark CXCR6+ ILC1s as potential memory cells of pregnancy.
Data were aligned to GRCm38 mouse genome (Ensembl Release 84) with TopHat2 (v2.1.1, using bowtie2 v2.2.9) with a double map strategy. Alignments and QC were processed using custom ClusterFlow (v0.5dev) pipelines and assessed using MultiQC (0.9.dev0). Gene quantification was determined with HTSeq-Counts (v0.6.1p1). Additional quality control was performed with feature counts (v 1.5.0-p2), qualimap (v2.2) and preseq (v2.0.0). Differential gene expression was performed with DESeq2 package (v1.16.0, R v3.4.2) and with the same package read counts were normalised on the estimated size factors.
| Resource | URL |
|---|---|
| GRCm38 | Link |
| FastQC | Link |
| Trim_galore | Link |
| TopHat2 | DOI |
| HTSeq-counts | DOI |
| Feature_counts | DOI |
| Qualimap | DOI |
| ClusterFlow | DOI |
| MultiQC | DOI |
A custom module for TopHat2 double map is provided in this repository, and can be run, by copying it into the modules directory of a ClusterFlow installation. Using just the HTSeq-Counts gene count tables, figures in the table below can be reproduced with the R script provided in this repository.
All files are provides in this repository with the exception of the GFF file for the mouse reference genome. The GFF file can be downloaded (ftp://ftp.ensembl.org/pub/release-84/gtf/mus_musculus/Mus_musculus.GRCm38.84.gtf.gz) and is required for the calculation of transcript lengths in the R script provided. Once downloaded the GFF file can be uncompressed using the command:
gunzip Mus_musculus.GRCm38.84.gtf.gz
The provided R script assumes the script is placed in a directory containing a subdirectory (called HTSeq_Counts) with all the htseq-counts files (one per sample). The script can be run interactively in R-studio or as a batch using Rscript. Note that some of the figures in the manuscript have had some labels and axes manually edited so may differ slightly from those created by the script provided.
Note: Differential gene expression analysis was performed with DESeq2 (v1.16.0), before the introduction of lfcShrink in version 1.18.0. If the latest version of DESeq2 is used to recreate this analysis we have included extra code in the R script to apply lfcShrink if the version is >= 1.18.0. However as the shrinkage methods are different the results will not be identical.
| Figure | Output Filename | Description |
|---|---|---|
| 1B | 2018-Filipovic-Colucci_Fig.1B.pdf | Bubble Time Course Plot (main) |
| 1B inset | 2018-Filipovic-Colucci_Fig.1B.inset.pdf | Bubble Time Course Plot (inset) |
| S1 | 2018-Filipovic-Colucci_SuppFig.S1.pdf | Bubble Time Course Plot with individual points (main) |
| S1 inset | 2018-Filipovic-Colucci_SuppFig.S1.inset.pdf | Bubble Time Course Plot with individual points (inset) |
| 2A | 2018-Filipovic-Colucci_Fig.2A.pdf | pHeatmap (top DEGs, all samples) |
| 2B | 2018-Filipovic-Colucci_Fig.2B.pdf | PCA all samples |
| 2C p1 | 2018-Filipovic-Colucci_Fig.2Cp1.pdf | Custom Expression Plot (Panel 1) |
| 2C p2 | 2018-Filipovic-Colucci_Fig.2Cp2.pdf | Custom Expression Plot (Panel 2) |
| 2D | 2018-Filipovic-Colucci_Fig.2D.pdf | Custom Expression Plot (Panel 3) |
| S2A | 2018-Filipovic-Colucci_SuppFig.S2A.pdf | PCA PC1 Explained |
| S2B | 2018-Filipovic-Colucci_SuppFig.S2B.pdf | PCA PC2 Explained |
| 3A | 2018-Filipovic-Colucci_Fig.3A.pdf | UpSetR Plot |
| 3B | 2018-Filipovic-Colucci_Fig.3B.pdf | Custom pHeatMap |
| 3C | 2018-Filipovic-Colucci_Fig.3C.pdf | Custom pHeatMap |
| 3D | 2018-Filipovic-Colucci_Fig.3D.pdf | Custom pHeatMap |
| 3E | 2018-Filipovic-Colucci_Fig.3E.pdf | Custom pHeatMap |
| 3F | 2018-Filipovic-Colucci_Fig.3F.pdf | Custom pHeatMap |
| S3A | 2018-Filipovic-Colucci_SuppFig.S3A.pdf | MA Plot |
| 4A | 2018-Filipovic-Colucci_Fig.4Ap1.pdf | Custom Expression Plot |
| 4F p1 | 2018-Filipovic-Colucci_Fig.4Fp1.pdf | Custom Expression Plot (Panel 1) |
| 4F p2 | 2018-Filipovic-Colucci_Fig.4Fp2.pdf | Custom Expression Plot (Panel 2) |
| 4F p3 | 2018-Filipovic-Colucci_Fig.4Fp3.pdf | Custom Expression Plot (Panel 3) |
| S4A | 2018-Filipovic-Colucci_Fig.S4A.pdf | DeconRNASeq Immune Cell Proportion Estimates |
| S4B | 2018-Filipovic-Colucci_Fig.S4B.pdf | DeconRNASeq Immune Cell Proportion Estimates |
It is possible to estimate the proportions of specific immune cell types from bulk RNA-Seq using reference data generated from known proportions of the cell types of interest (Chen et al, 2017). Methods such as DeconRNASeq (Gong & Szustakowski, 2013) take these tables of known cell proportions, defined by gene expression profiles, and use them to deconvolve bulk to estimate cell proportions with in each of the sequences samples.
Gong, T., & Szustakowski, J. D. (2013). DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics, 29(8), 1083–1085. 10.1093/bioinformatics/btt090
Chen, Z., Huang, A., Sun, J., Jiang, T., Qin, F. X.-F., & Wu, A. (2017). Inference of immune cell composition on the expression profiles of mouse tissue. Scientific Reports, 7, 40508. 10.1038/srep40508
UpSetR is an alternative for plotting sets of data to visualise overlaps as a more intuitive replacement for Euler/Venn Diagrams.
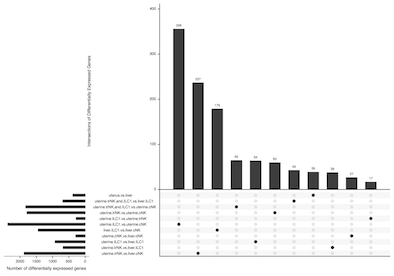
Conway, J. R., Lex, A., & Gehlenborg, N. (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics, 33(18), 2938–2940
10.1093/bioinformatics/btx364
Comma separated value (csv) files are generated in the Rscript to identify the intersections of genes for each of the comparisons made.
| Description | Filename |
|---|---|
| Bubble Time Course Plot table | 180410_all_points_uterus.txt |
| Bubble Time Course Plot inset table | 180410_nBF.txt |
| DeconRNASeq Immune Cell Proportions Table | 10.1038/srep40508 |
Details for the R version and packages used to create all figures
R version 3.4.2 (2017-09-28)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: OS X El Capitan 10.11.6
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] gtools_3.5.0 rtracklayer_1.38.3 UpSetR_1.3.3 scales_0.5.0
[5] plyr_1.8.4 reshape2_1.4.3 ggrepel_0.7.0 pheatmap_1.0.8
[9] RColorBrewer_1.1-2 DESeq2_1.18.1 SummarizedExperiment_1.8.1 DelayedArray_0.4.1
[13] matrixStats_0.53.1 Biobase_2.38.0 GenomicRanges_1.30.3 GenomeInfoDb_1.14.0
[17] IRanges_2.12.0 S4Vectors_0.16.0 BiocGenerics_0.24.0 genefilter_1.60.0
[21] cowplot_0.9.2 ggplot2_2.2.1 biomaRt_2.34.2
loaded via a namespace (and not attached):
[1] httr_1.3.1 bit64_0.9-7 splines_3.4.2 Formula_1.2-2
[5] assertthat_0.2.0 latticeExtra_0.6-28 blob_1.1.0 BSgenome_1.46.0
[9] Rsamtools_1.30.0 GenomeInfoDbData_1.0.0 yaml_2.1.17 progress_1.1.2
[13] pillar_1.2.1 RSQLite_2.0 backports_1.1.2 lattice_0.20-35
[17] digest_0.6.15 XVector_0.18.0 checkmate_1.8.5 colorspace_1.3-2
[21] htmltools_0.3.6 Matrix_1.2-12 XML_3.98-1.10 zlibbioc_1.24.0
[25] xtable_1.8-2 BiocParallel_1.12.0 htmlTable_1.11.2 tibble_1.4.2
[29] annotate_1.56.1 nnet_7.3-12 lazyeval_0.2.1 survival_2.41-3
[33] magrittr_1.5 memoise_1.1.0 MASS_7.3-49 foreign_0.8-69
[37] BiocInstaller_1.28.0 tools_3.4.2 data.table_1.10.4-3 prettyunits_1.0.2
[41] stringr_1.3.0 munsell_0.4.3 locfit_1.5-9.1 cluster_2.0.6
[45] Biostrings_2.46.0 AnnotationDbi_1.40.0 compiler_3.4.2 rlang_0.2.0
[49] grid_3.4.2 RCurl_1.95-4.10 rstudioapi_0.7 htmlwidgets_1.0
[53] labeling_0.3 bitops_1.0-6 base64enc_0.1-3 gtable_0.2.0
[57] curl_3.1 DBI_0.7 R6_2.2.2 GenomicAlignments_1.14.1
[61] gridExtra_2.3 knitr_1.20 bit_1.1-12 Hmisc_4.1-1
[65] stringi_1.1.6 Rcpp_0.12.15 geneplotter_1.56.0 rpart_4.1-13
[69] acepack_1.4.1
| sample Name | cell | tissue |
|---|---|---|
| SLX-9343.01 | cNK | uterus |
| SLX-9343.02 | trNK | uterus |
| SLX-9343.04 | ILC1 | liver |
| SLX-9343.05 | cNK | liver |
| SLX-9343.06 | cNK | uterus |
| SLX-9343.07 | trNK | uterus |
| SLX-9343.08 | ILC1 | uterus |
| SLX-9343.09 | ILC1 | liver |
| SLX-9343.10 | cNK | liver |
| SLX-9343.12 | trNK | uterus |
| SLX-9343.13 | ILC1 | uterus |
| SLX-9343.14 | ILC1 | liver |
| SLX-9343.15 | cNK | liver |
| Description | URL |
|---|---|
| Publication | [Nature Communications, 9, 4492] [DOI] [bioRxiv] [DOI] |
| Raw Data | ArrayExpress EMBL-EBI E-MTAB-6812 |
| Colucci Group | Colucci group website |
| CTR Bioinformatics | CTR-BFX |
Contact rsh46 -at- cam.ac.uk for bioinformatics related queries
