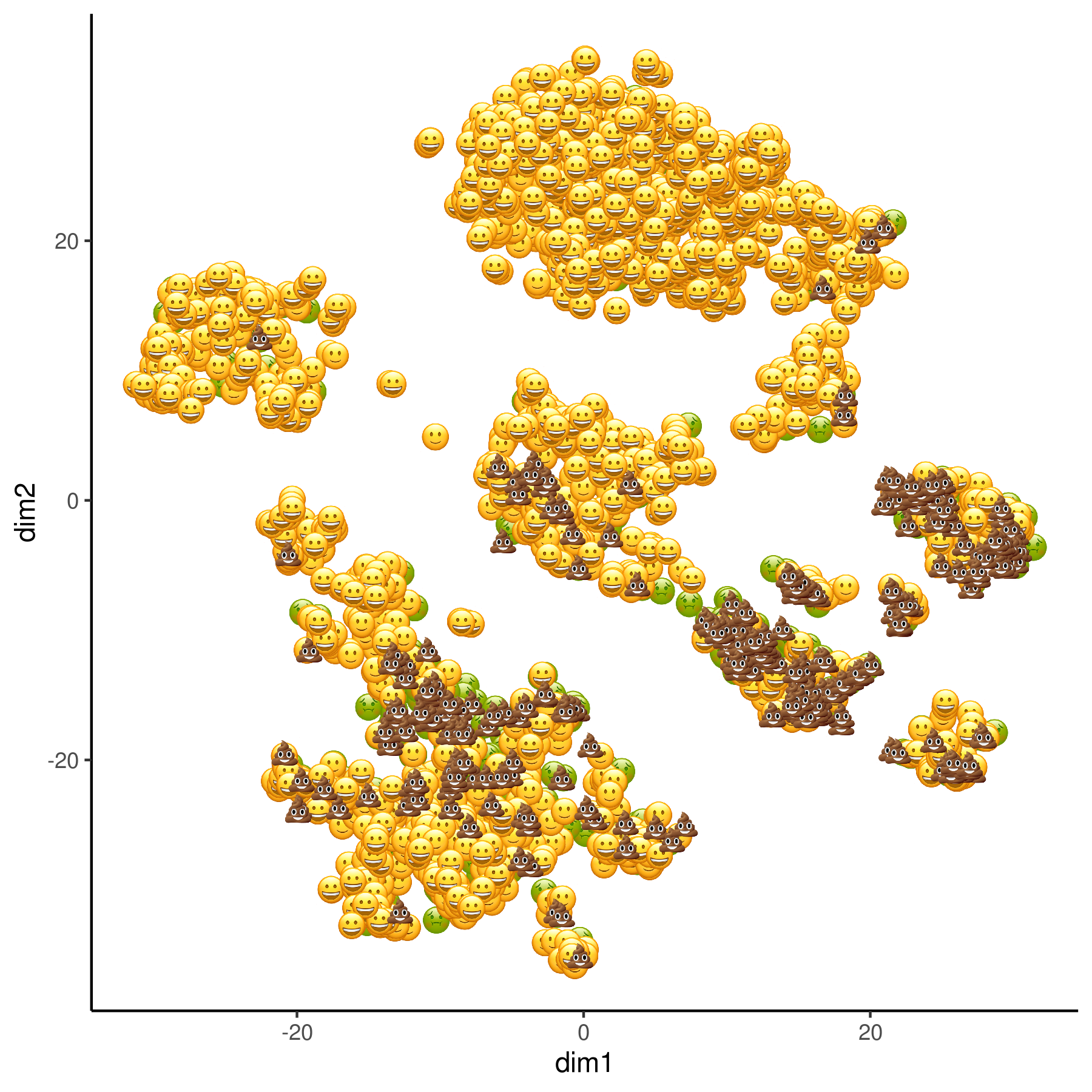
scrappy provides an easy way to visualize the quality of scRNA single-cells.
You can install the development version with:
# install.packages("remotes")
remotes::install_github("darlanminussi/scrappy")scrappy uses the percentage of mitochondrial gene expression to classify the cells as:
😄: Great Quality.
🙂: Good quality.
🤢: OK quality.
💩: Low quality.
scrappy can be used on SingleCellExperiment objects as well as Seurat objects.
sce
#> class: SingleCellExperiment
#> dim: 20006 3005
#> metadata(0):
#> assays(2): counts logcounts
#> rownames(20006): Tspan12 Tshz1 ... mt-Rnr1 mt-Nd4l
#> rowData names(1): featureType
#> colnames(3005): 1772071015_C02 1772071017_G12 ... 1772066098_A12
#> 1772058148_F03
#> colData names(26): tissue group # ... altexps_repeat_percent total
#> reducedDimNames(2): PCA TSNE
#> spikeNames(0):
#> altExpNames(2): ERCC repeatlibrary(scrappy)
scrappyPlot(sce, "TSNE")PS: If you want to learn about QC metrics and how to filter your single-cell datasets follow this link.