fastq-profiler is a command line utility for keeping track of fastqs and process and store information associated with them. fastq-profiler generates summary statistics from processed fastq files and stores the data using the files md5sum as an identifier in Google Datastore.
I chose Google Datastore because it is centralized - allowing you to profile fastqs locally, or within cluster environments and elsewhere in parallel without having to track/combine files. fastq-profiler also integrates with FastQC and can be used to output fastq statistics for analysis. Importantly, when duplicates are identified, fastq-profiler keeps track of both locations, allowing you to identify manage duplicate files if necessary.
pip install https://github.com/danielecook/fastq-profiler/archive/v0.0.4.tar.gz
- Setup an account with google cloud.
- Authorize Google Cloud using the gcloud SDK:
gcloud auth login
- Set your project and default Google Datastore "kind" using:
fq set <project> <kind>
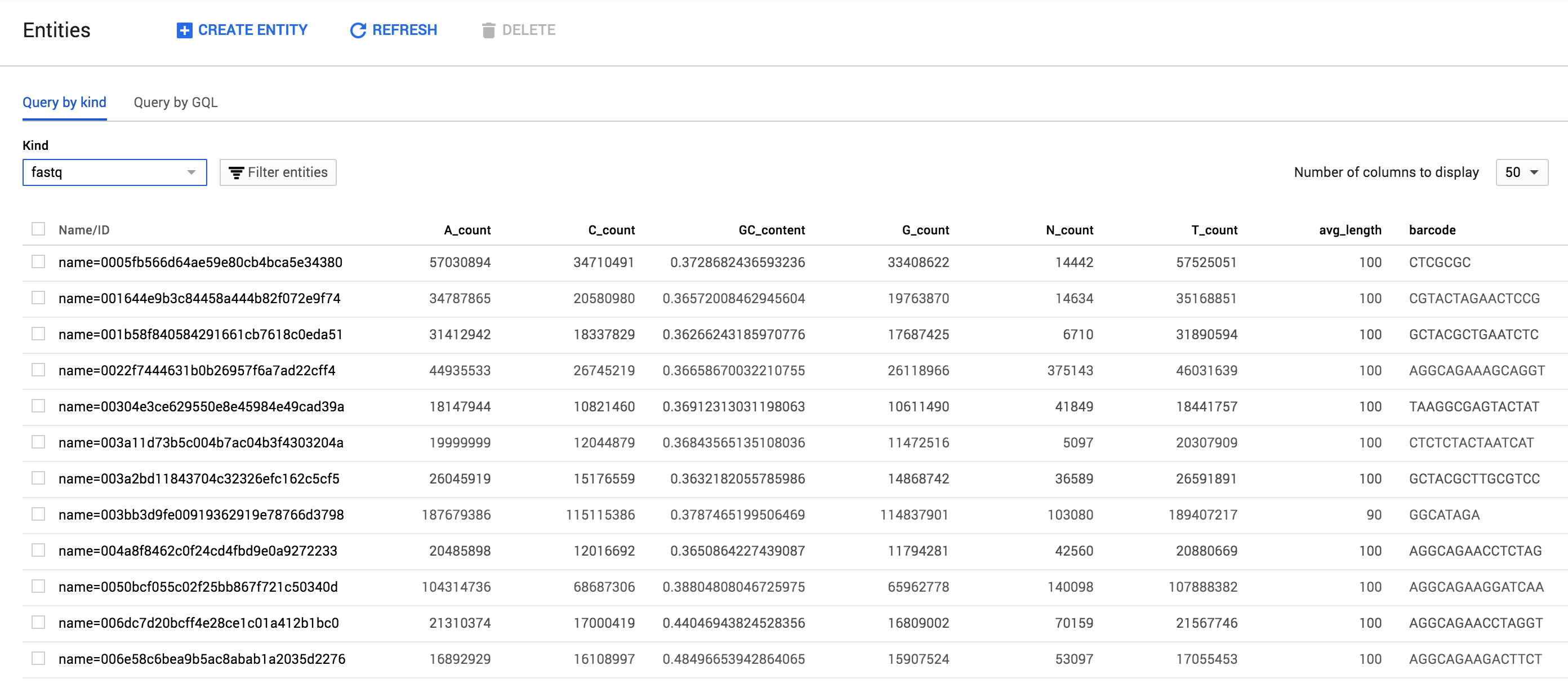
fastq-profiler generates a hash of every fastq submitted and uses it to track and store data about fastqs. The end result looks like this within the browsable Google Datastore interface:
The following pieces of information about fastqs (properties) are stored:
Array Elements
The three properties below are stored as arrays. Elements within those arrays correspond with one another (e.g. [1,2] ~ [A,B])
hostname- HostnamebasenameFastq basenamefilename- Absolute path of Fastq (in every location identified)
Singlular properties
md5sumHash of the filelocations_count- Count of identified locations.date_createdEarliest identified date createdflowcell_laneFlowcell lanefilesize- Filesize in byteshfilesize- Filesize in human readable formtotal_reads- Read count[ATCGN]_count- Base countsGC_content- GC contentmin_length- Minimum read lengthavg_length- Average read lengthmax_length- maximum read length
Additional fields included if applicable
instrumentInstrument name if availableflowcell_laneflowcell_numberrun_idpair1/2 for paired end sequencing.barcodeIndex/barcode of read for pooled sequencingcontrol_bits
Illumina Filename
If the filename follows the Illumina filename conventions, these items will be parsed out as well:
illumina_filename_sampleillumina_filename_barcode_sequenceORillumina_filename_sample_numberillumina_filename_laneillumina_filename_readillumina_filename_set_number
For example:
EA-CFB-2-421_S1_L001_R1_001.fastq.gz would be parsed into:
illumina_filename_sample= EA-CFB-2-421illumina_filename_sample_number= S1illumina_filename_lane= L001illumina_filename_read= R1illumina_filename_set_number1
fq profile creates a .checksum file in every directory containing fastqs that it is run on. The .checksum file is used as a cache of file hashes to make retrieval of data easier and help with file tracking.
FastQC Stats
fastq-profiler can optionally store FastQC results in datastore as well, and these results can be output as one file, enabling easy aggregation of fastqc results. To incorporate FastQC data, be sure to use the --fastqc flag:
fq profile --fastqc <fq>
fastqc-profiler will store the following data in Google Datastore as unindexed properties:
- per_base_sequence_quality
- per_tile_sequence_quality
- per_sequence_quality_scores
- per_base_sequence_content
- per_sequence_gc_content
- per_base_n_content
- sequence_length_distribution
- sequence_duplication_levels
- overrepresented_sequences
- adapter_content
- kmer_content
Set your project and kind:
fq set <project> <kind>
Set <project> to your google cloud project name. Set kind to the name of the kind you want to store fastq data in within Google Datastore.
fq set 'my-google-cloud-project' 'fastq-set'
Profile a fastq
The profile command is designed to be run on any/all fastqs you have, even if they are duplicates. When duplicate files are identified, the path and filenames are both stored under the same entry. However, because they have the same file hash, fastq-profiler does not repeat profiling or fastqc operations.
fq profile [options] <fq>...
Run fq profile on multiple fastqs
fq profile myseq1.fq.gz myseq2.fq.gz myseq3.fq.gz
Run fq profile on an entire directory
You can use a * wildcard:
fq profile *.fq.gz
Read files from stdin
find . -name *.gz | egrep "(fastq|fq)" - | fq profile -
The .description file is intended to specify data that should be attached to every fastq within a folder. .description files are located in the directory containing fastqs that will be processed. For example:
├── 20150505_fastq_files
│ ├── .description
│ ├── sample_001_R01.fq.gz
│ ├── sample_001_R02.fq.gz
|...|
│ ├── sample_300_R01.fq.gz
│ └── sample_300_R02.fq.gz
The format of the description file is key, value pairs separated by a :. For example:
species: C. elegans
source: ftp_site
sequencing_center: UChicago
seq_folder: 150406_D00422_0191_BHBDWCADXX-EA-CB12
sequencing_type: DNA
date_submitted: date-2015-03-05
date_sequenced: date-2015-04-06
lab: Andersen Lab
description: DNA sequencing of new wild isolates!
Every datastore entity corresponding to a fastq in the folder will have these data attached when you run fq profile.
If you want to store data specific to the fastq file, for the sequencing library or other properties, you can use a .fqdata file. The .fqdata file functions similar to the description file except that you must specify the fastq using its basename (e.g. /fq_set/myfastq.fq.gz would be myfastq.fq.gz) or md5sum hash.
By the way, you can use both .description and .fqdata files!
├── 20150505_fastq_files
│ ├── .fqdata
│ ├── .description
│ ├── sample_001_R01.fq.gz
│ ├── sample_001_R02.fq.gz
|...|
│ ├── sample_300_R01.fq.gz
│ └── sample_300_R02.fq.gz
The .fqdata file is tab-separated. For example:
#file Library prepared_by dna_prep_kit
sample_001_R01.fq.gz LIB1 Robyn A
sample_001_R02.fq.gz LIB2 Robyn B
sample_002_R01.fq.gz LIB3 Mostafa C
…
The table looks like this:
| file | Library | prepared_by | dna_prep_kit |
|---|---|---|---|
| sample_001_R01.fq.gz | LIB1 | Robyn | A |
| sample_001_R02.fq.gz | LIB2 | Robyn | B |
| sample_002_R01.fq.gz | LIB3 | Mostafa | C |
| ... | ... | ... | ... |
To specify dates use the date- prefix and use YYYY-MM-DD. For example, date-2015-03-05.
Once you have profiled fastqs, you can fetch data associated with them using the fetch command:
fq profile fetch myseq1.fq.gz myseq2.fq.gz
The fetch command omits FastQC data tables stored in Datastore.
Output
Output is in JSON format.
{
"A_count": 94645601,
"C_count": 59898634,
...
"barcode": "GAATCTC",
"basename": [
"EA-B07_GAATCTC_L001_R2_001.fastq.gz"
],
"bases": 309854148,
"basic_statistics": "pass",
"control_bits": 0,
"cum_length": 309858000,
"date_created": "2015-03-02T10:24:08+00:00",
...
"filename": [
"/Volumes/PortusTutus/Project_EA-DS/Sample_EA-B07/EA-B07_GAATCTC_L001_R2_001.fastq.gz"
],
"filesize": 245108588,
"filtered": "N",
"flowcell_id": "HGMN3ADXX",
"flowcell_lane": 1,
"fq_profile_count": 1,
"hfilesize": "233 MiB",
"hostname": [
"dancook"
],
...
"sequence_length_distribution": "pass",
"total_reads": "3098580",
"unique_reads": "2002529"
}
Alternatively, you can dump fastq data stored that is stored in the kind you set with fq set:
fq dump
The command above will dump all fastq data in JSON format. Data tables saved by FastQC are ommited.
fastq-profiler can store FastQC data, enabling easy aggregation of fastqc results. To use, you must profile fastqs with the --fastqc flag. For example:
fq profile --fastqc <fq>
Then, you can output data using:
fq fastq-dump <fastqc-group> [<fq>...]
Where is one of:
- per_base_sequence_quality
- per_tile_sequence_quality
- per_sequence_quality_scores
- per_base_sequence_content
- per_sequence_gc_content
- per_base_n_content
- sequence_length_distribution
- sequence_duplication_levels
- overrepresented_sequences
- adapter_content
- kmer_content
For example:
fq fastqc-dump per_base_sequence_content *.fq.gz
Will output all fastqc data in one file among files matching the *.fq.gz wildcard.
Output
filename base G A T C
NIC1_130123_I186_FCC1GJUACXX_L2_CHKPEI13010005_1.fq.gz 1 39.08 25.0 12.36 23.56
NIC1_130123_I186_FCC1GJUACXX_L2_CHKPEI13010005_1.fq.gz 2 15.952184666117065 22.176422093981863 41.13767518549052 20.733718054410552
NIC1_130123_I186_FCC1GJUACXX_L2_CHKPEI13010005_1.fq.gz 3 17.0 28.360000000000003 33.0 21.64
...
t_IndexQX1791_2.fq.gz 84-85 18.82 30.54 32.28 18.360000000000003
t_IndexQX1791_2.fq.gz 86-87 18.58 30.159999999999997 33.18 18.08
t_IndexQX1791_2.fq.gz 88-89 18.44 29.439999999999998 31.96 20.16
t_IndexQX1791_2.fq.gz 90 18.6 30.48 31.28 19.64
If you specify a list of fastqs, only fastqc results for those files will be dumped. If you leave <fq>... empty, all fastqc data for the <fastqc-group> specified will be output under the set kind. Note: If a fastq is duplicated, it will only output the first filename that has been stored.
--fastqc-threads
Use --fastqc-threads to speed up fastqc.
fq profile --fastqc --fastqc-threads 8 <fq>
If you use fq dump or fq fetch you can import data into R using jsonlite:
library(jsonlite)
fromJSON("out.json")
--kv=<k:v> can be used to store custom data.
Use the date- prefix to store a date. Other data types are automatically inferred. You can alternatively use a .description file.
fq profile --kv=date_sequenced:date-20160610,sequencing_center:UChicago *.fq.gz
In the example above, a 'date_sequenced' property will be added to the fastq entity in google datastore.
--verbose - Provide additional information on what is going on under the hood.