Refer to the issue, this document will reproduce functional analysis using DOSE and clusterProfiler packages.
The paper and supplemental files are located at paper folder.
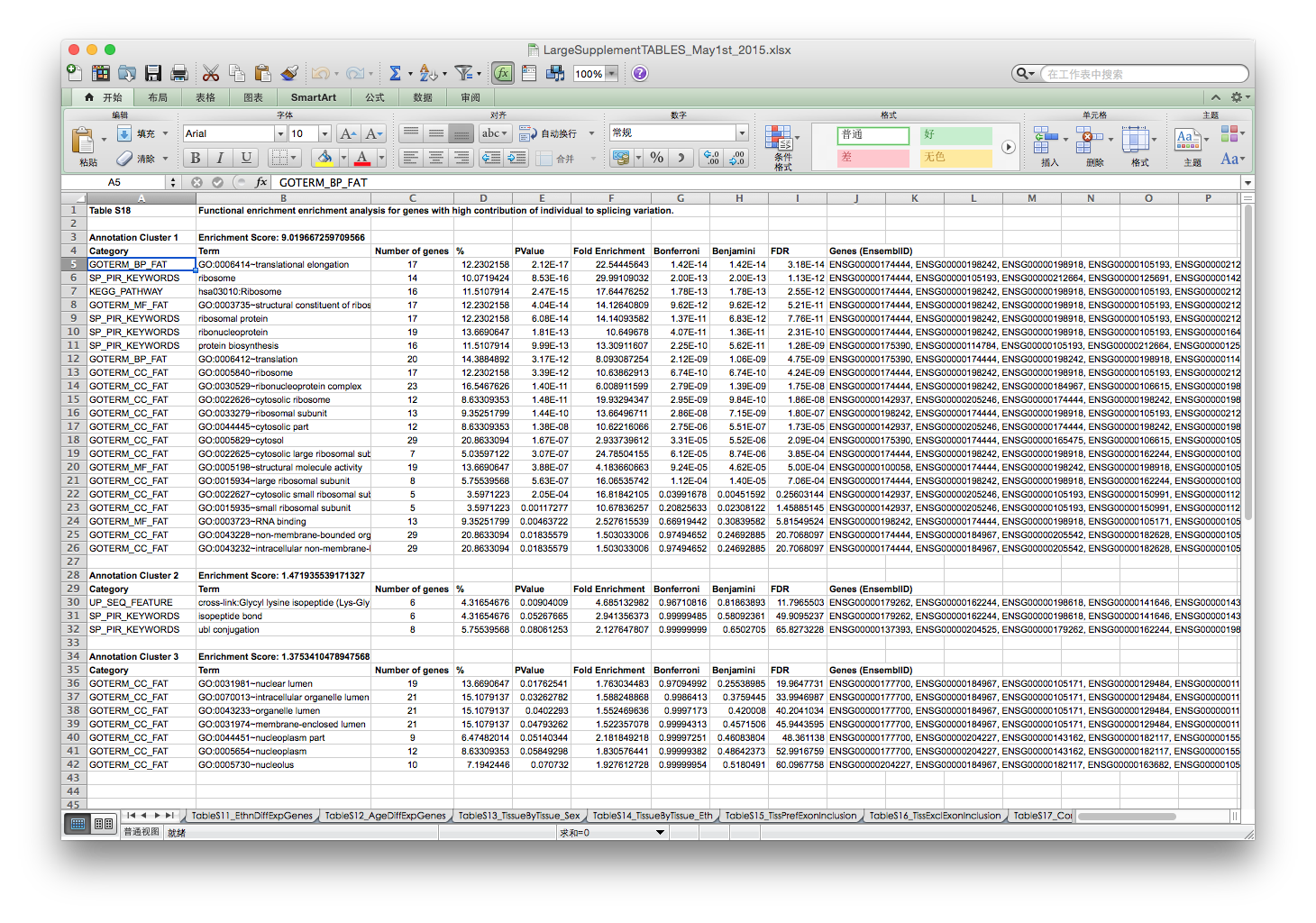
In page 63 of supplemental file, the authors mentioned that genes with high contribution of individuals to splicing variation were used in GO enrichment analysis.
The Individual and Tissue contribution to variation of splicing was stored in supplemental table 17, which was exported as a csv file.
require(magrittr)
require(DOSE)
require(RDAVIDWebService)
require(clusterProfiler)
table17 <- read.csv("paper/tableS17.csv")
gene <- with(table17, gene_id[ Individuals > quantile(Individuals, 0.98)])
gene %<>% as.character %>% gsub("\\.\\d+", "", .)
head(gene)
## [1] "ENSG00000002586" "ENSG00000011007" "ENSG00000014164" "ENSG00000042753"
## [5] "ENSG00000051620" "ENSG00000055130"
length(gene)
## [1] 212
The authors did not mention how they define high contribution, here I use the top 2% of genes and get 212 genes with 168 genes that could be mapped in the DAVID database. Slightly large than the number (139) reported in supplemental file.
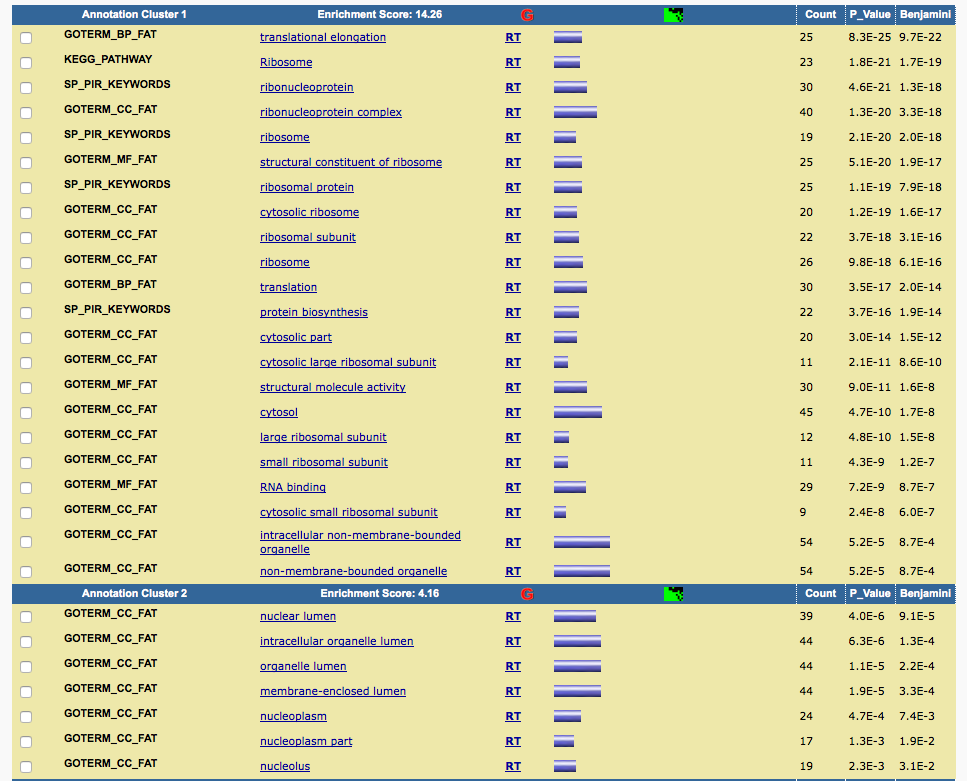
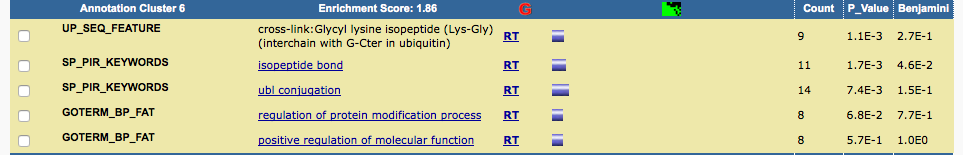
The GO enrichment result reported in supplemental table 18 is:
I pasted the genes into DAVID and got similar results.
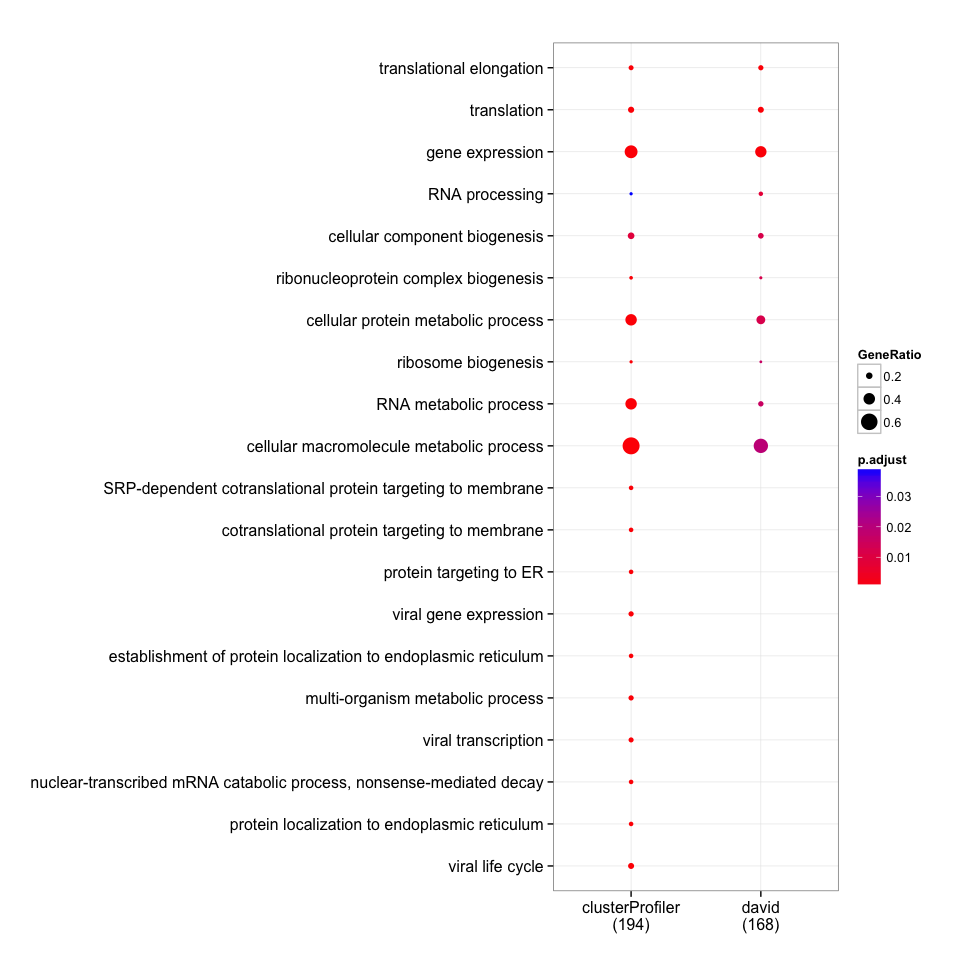
Although the gene list I selected here is slighly different from the one author selected (which we don't know), it can reproduce the results reported in the paper.
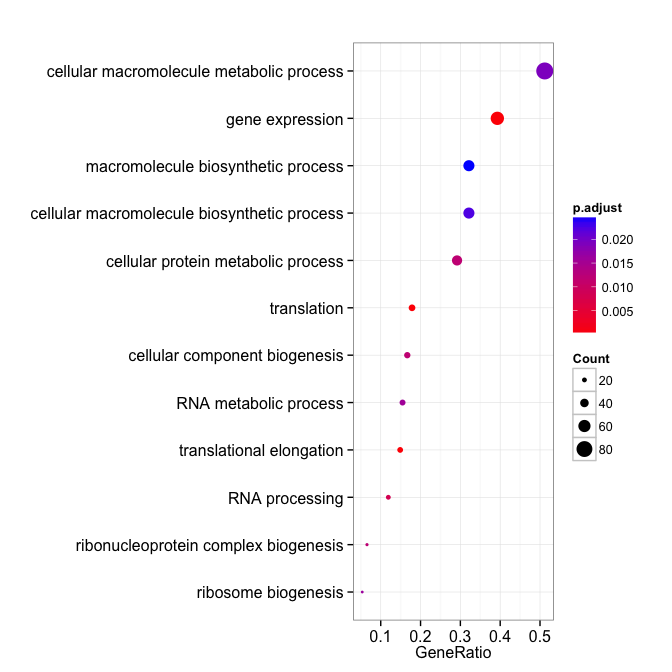
david_bp <- enrichDAVID(gene, idType="ENSEMBL_GENE_ID", annotation="GOTERM_BP_ALL", qvalueCutoff=0.05, david.user="gcyu@connect.hku.hk")
summary(david_bp)[, -8]
## ID Description
## GO:0006414 GO:0006414 translational elongation
## GO:0006412 GO:0006412 translation
## GO:0010467 GO:0010467 gene expression
## GO:0006396 GO:0006396 RNA processing
## GO:0044085 GO:0044085 cellular component biogenesis
## GO:0022613 GO:0022613 ribonucleoprotein complex biogenesis
## GO:0044267 GO:0044267 cellular protein metabolic process
## GO:0042254 GO:0042254 ribosome biogenesis
## GO:0016070 GO:0016070 RNA metabolic process
## GO:0044260 GO:0044260 cellular macromolecule metabolic process
## GO:0034645 GO:0034645 cellular macromolecule biosynthetic process
## GO:0009059 GO:0009059 macromolecule biosynthetic process
## GO:0006397 GO:0006397 mRNA processing
## GO:0032268 GO:0032268 regulation of cellular protein metabolic process
## GeneRatio BgRatio pvalue p.adjust qvalue
## GO:0006414 25/168 101/14116 4.833921e-25 6.298599e-22 9.718725e-23
## GO:0006412 30/168 331/14116 1.916789e-17 1.248788e-14 1.926877e-15
## GO:0010467 66/168 2999/14116 1.313901e-07 5.706548e-05 8.805443e-06
## GO:0006396 20/168 547/14116 2.561454e-05 8.309328e-03 1.287468e-03
## GO:0044085 28/168 1001/14116 4.549872e-05 1.178722e-02 1.829527e-03
## GO:0022613 11/168 180/14116 5.566722e-05 1.201662e-02 1.844751e-03
## GO:0044267 49/168 2355/14116 6.422825e-05 1.188482e-02 1.844751e-03
## GO:0042254 9/168 122/14116 9.956471e-05 1.608661e-02 2.434515e-03
## GO:0016070 26/168 938/14116 1.089796e-04 1.565485e-02 2.434515e-03
## GO:0044260 86/168 5214/14116 1.478656e-04 1.908386e-02 2.972876e-03
## GO:0034645 54/168 2812/14116 1.896884e-04 2.222098e-02 3.467032e-03
## GO:0009059 54/168 2832/14116 2.282688e-04 2.448429e-02 3.824503e-03
## GO:0006397 13/168 321/14116 4.392293e-04 4.307854e-02 6.792939e-03
## GO:0032268 16/168 474/14116 5.095183e-04 4.632629e-02 7.317143e-03
## Count
## GO:0006414 25
## GO:0006412 30
## GO:0010467 66
## GO:0006396 20
## GO:0044085 28
## GO:0022613 11
## GO:0044267 49
## GO:0042254 9
## GO:0016070 26
## GO:0044260 86
## GO:0034645 54
## GO:0009059 54
## GO:0006397 13
## GO:0032268 16
dotplot(david_bp, showCategory=12)
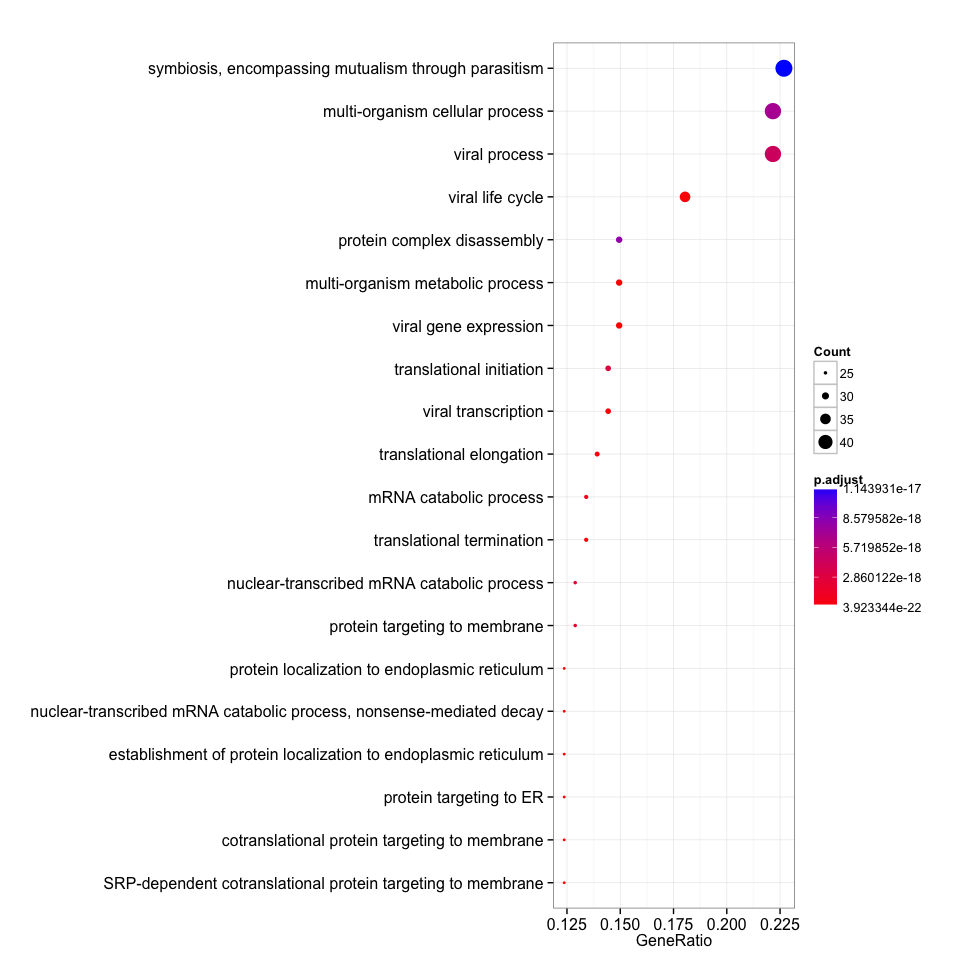
eg=bitr(gene, "ENSEMBL", "ENTREZID", "org.Hs.eg.db")[, "ENTREZID"]
clusterProfiler_bp <- enrichGO(eg, ont="BP")
dotplot(clusterProfiler_bp, showCategory=20)
merge_result(list(david=david_bp, clusterProfiler=clusterProfiler_bp)) %>%
plot(., showCategory=10)
date()
## [1] "Wed Jul 15 21:15:17 2015"
sessionInfo()
## R version 3.2.1 (2015-06-18)
## Platform: x86_64-apple-darwin14.3.0 (64-bit)
## Running under: OS X 10.10.4 (Yosemite)
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats4 parallel stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] clusterProfiler_2.3.6 RDAVIDWebService_1.6.0 ggplot2_1.0.1
## [4] GOstats_2.34.0 Category_2.34.2 GO.db_3.1.2
## [7] AnnotationDbi_1.30.1 GenomeInfoDb_1.4.1 IRanges_2.2.5
## [10] S4Vectors_0.6.1 Matrix_1.2-2 Biobase_2.28.0
## [13] BiocGenerics_0.14.0 graph_1.46.0 DOSE_2.7.9
## [16] RSQLite_1.0.0 DBI_0.3.1 rmarkdown_0.7
## [19] roxygen2_4.1.1 magrittr_1.5 BiocInstaller_1.18.3
##
## loaded via a namespace (and not attached):
## [1] KEGGREST_1.8.0 qvalue_2.0.0 genefilter_1.50.0
## [4] reshape2_1.4.1 splines_3.2.1 rJava_0.9-6
## [7] lattice_0.20-31 colorspace_1.2-6 htmltools_0.2.6
## [10] XML_3.98-1.3 RBGL_1.44.0 survival_2.38-3
## [13] topGO_2.20.0 plyr_1.8.3 stringr_1.0.0
## [16] zlibbioc_1.14.0 Biostrings_2.36.1 munsell_0.4.2
## [19] GOSemSim_1.27.4 gtable_0.1.2 evaluate_0.7
## [22] labeling_0.3 knitr_1.10.5 SparseM_1.6
## [25] GSEABase_1.30.2 proto_0.3-10 Rcpp_0.11.6
## [28] xtable_1.7-4 scales_0.2.5 formatR_1.2
## [31] DO.db_2.9 annotate_1.46.1 XVector_0.8.0
## [34] png_0.1-7 digest_0.6.8 stringi_0.4-1
## [37] grid_3.2.1 tools_3.2.1 MASS_7.3-42
## [40] httr_1.0.0 AnnotationForge_1.10.1 R6_2.1.0
## [43] igraph_1.0.1