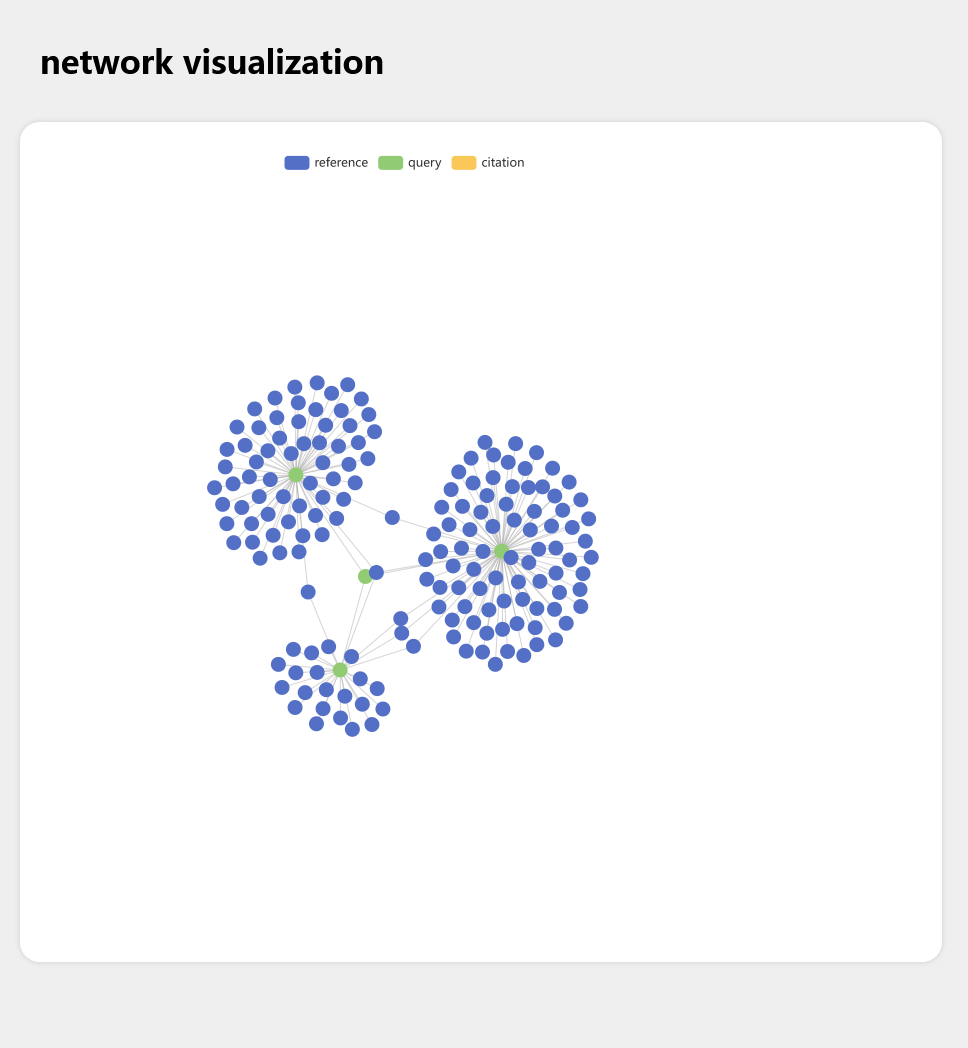
This project is aimed at make a citation map visualizing tool, which can visualize the reference/citation relations of articles on PubMed dataset.
./
|- app.py
|- get_result_in_table_formation.py
|- citation_map.json
|- example.PMID.txt
|- index.html
|- README.md
|
\---img
\--- example.png
First, we define three kinds of articles:
- Query articles: These articles are user input, and we will use these "Query" to query PubMed dataset.
- Reference articles: These articles are "Query"'s reference, in other words, "Query" articles cited these articles.
- Citation articles: These articles are "Query"'s citation, in other words, "Query" articles are cited by these articles.
Right now we can extract "Reference" from Pubmed, and in the future we will extract "Citation" articles as well
When user input a list of articles' PMID(this is important! the input should be PMID), we will make a query to PubMed by its web API, and get a report in XML format.
Then we will parse this XML report and generate a .json file, which contains information of graph nodes(article) and graph edges(citation relation).
Finally, we will use Echarts to visualize this citation map graph. The final report is in .html format , and you need to open your browser to read and interact with it.
All requirements:
python(>=3.6)with The Python Standard Libraryrequests(>=2.27).
This program don't need installation; just download these file is enough.
The build-in help text is:
usage: app.py [-h] [-f FILE | -l LIST] [-m MAX] [-p PORT]
optional arguments:
-h, --help show this help message and exit
-f FILE, --file FILE PMID list file.
-l LIST, --list LIST PMID list(comma seperate).
-m MAX, --max MAX Max limitation of query article number. Default is 10.
-p PORT, --port PORT Network port of the report server. Default is using
random number.
To use this program, first you need to prepare a list of PMIDs. You can storage them into a seperate text file, or just copy to the clip-board. Then you open a terminal and switch working directory to where you download this project. You have two method to launch this program:
This method will load pmids from a seperate text file. in that text file, each id occupies a separate row, and there should no blank before pmid.
Example:
24651067
32286628
32184769
31606751
PMID list file also can contain comment text, but comment text should start with a "#" character.
Example:
# These articles are some studies about CRISPR-Cas9 technology
24651067 # Hum Mol Genet. 2014 Sep 15;23(R1):R40-6.
32286628 # Nucleic Acids Res. 2020 May 21;48(9):4698-4708.
32184769 # Front Microbiol. 2020 Feb 28;11:282.
31606751 # Hum Genet. 2019 Dec;138(11-12):1217-1225
The above PMID list file names example.PMID.txt, and is storaged in current directory. Then use the following command to launch the program:
python ./app.py -f ./example.PMID.txtThe program will load PMID list and get citation relation from PubMed web API. You will see the output as below:
~$ python ./app.py -f ./example.PMID.txt
Max limitation of query article number is 10.
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=pubmed&id=24651067,32286628,32184769,31606751
Default port number will be set.
The report is serving at port 15501.
Please open the following URL to check report: http://localhost:15501
To stop web server, Press `Ctrl+C`
127.0.0.1 - - [12/Dec/2023 21:10:40] "GET / HTTP/1.1" 200 -
127.0.0.1 - - [12/Dec/2023 21:10:41] "GET /citation_map.json HTTP/1.1" 200 -
You can open your web browser and access the URL which is mentioned in the output.
This method will load pmids from command line. in this method, the argument -l will be used. You should provide a comma seperate-PMID list, like this:
python ./app.py -l 24651067,32286628,32184769,31606751The output is the same as above method.
I provide a stript file called get_result_in_table_formation.py, which is for converting json document into a readable tsv file.
Usage:
python get_result_in_table_formation.py
(with no parameter. The script will load citation_map.json file and write result into result_exported.tsv file.)
The output file result_exported.tsv is like this:
index count PMID title journal authorList DOI date PMC citation abstract category name
0 0 250.0 24651067 CRISPR/Cas9 for genome editing: progress, implications and challenges. Human molecular genetics ['Zhang Feng', 'Wen Yan', 'Guo Xiong']10.1093/hmg/ddu125 2015-05-11 Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) protein 9 system provides a robust and multiplexable genome editing tool, enabling researchers to precisely manipulate specific genomic elements, and facilitating the elucidation of target gene function in biology and diseases. CRISPR/Cas9 comprises of a nonspecific Cas9 nuclease and a set of programmable sequence-specific CRISPR RNA (crRNA), which can guide Cas9 to cleave DNA and generate double-strand breaks at target sites. Subsequent cellular DNA repair process leads to desired insertions, deletions or substitutions at target sites. The specificity of CRISPR/Cas9-mediated DNA cleavage requires target sequences matching crRNA and a protospacer adjacent motif locating at downstream of target sequences. Here, we review the molecular mechanism, applications and challenges of CRISPR/Cas9-mediated genome editing and clinical therapeutic potential of CRISPR/Cas9 in future. 1 Zhang Feng,2015
1 1 106.0 32286628 Machine learning predicts new anti-CRISPR proteins. Nucleic acids research ['Eitzinger Simon', 'Asif Amina', 'Watters Kyle E', 'Iavarone Anthony T', 'Knott Gavin J', 'Doudna Jennifer A', 'Minhas Fayyaz Ul Amir Afsar'] 10.1093/nar/gkaa219 2020-09-08 The increasing use of CRISPR-Cas9 in medicine, agriculture, and synthetic biology has accelerated the drive to discover new CRISPR-Cas inhibitors as potential mechanisms of control for gene editing applications. Many anti-CRISPRs have been found that inhibit the CRISPR-Cas adaptive immune system. However, comparing all currently known anti-CRISPRs does not reveal a shared set of properties for facile bioinformatic identification of new anti-CRISPR families. Here, we describe AcRanker, a machine learning based method to aid direct identification of new potential anti-CRISPRs using only protein sequence information. Using a training set of known anti-CRISPRs, we built a model based on XGBoost ranking. We then applied AcRanker to predict candidate anti-CRISPRs from predicted prophage regions within self-targeting bacterial genomes and discovered two previously unknown anti-CRISPRs: AcrllA20 (ML1) and AcrIIA21 (ML8). We show that AcrIIA20 strongly inhibits Streptococcus iniae Cas9 (SinCas9) and weakly inhibits Streptococcus pyogenes Cas9 (SpyCas9). We also show that AcrIIA21 inhibits SpyCas9, Streptococcus aureus Cas9 (SauCas9) and SinCas9 with low potency. The addition of AcRanker to the anti-CRISPR discovery toolkit allows researchers to directly rank potential anti-CRISPR candidate genes for increased speed in testing and validation of new anti-CRISPRs. A web server implementation for AcRanker is available online at http://acranker.pythonanywhere.com/. 1 Eitzinger Simon,2020
in which the count column indicates how many edges are there related to the current article. The articles in this tsv file is sorted by count value.
- Due to the limitation of PubMed web API, some articles' reference list cannot load correctly. Therefore, the report cannot display all citation relationships.
- Current version of this program only can display reference network. In the future we will support citation network.
- If input too much articles into program, the render on the web browser will be very slow. Recommend input is less than 10 articles. In the future we will support node pruning, which will remove some low citation/reference articles.